Virus (biology): Difference between revisions
imported>Nancy Sculerati MD No edit summary |
imported>Nancy Sculerati MD m (→Origins) |
||
| Line 28: | Line 28: | ||
==Origins== | ==Origins== | ||
The origins of modern viruses are not entirely clear, and there may not be a single mechanism of origin that can account for all viruses. As viruses do not [[fossil|fossilise]] well, [[Molecular biology|molecular techniques]] have | The origins of modern viruses are not entirely clear, and there may not be a single mechanism of origin that can account for all viruses. As viruses do not [[fossil|fossilise]] well, [[Molecular biology|molecular techniques]], rather than the geologic records, have provided the most useful means of hypothesising how they arose. Research in [[microfossil]] identification and molecular biology may yet discern fossil evidence dating to the [[Archean]] or [[Proterozoic]] eons. Two main hypotheses currently exist<ref>Prescott, L. (1993). Microbiology, Wm. C. Brown Publishers, ISBN 0-697-01372-3</ref>: | ||
*Small viruses with only a few genes may be runaway stretches of nucleic acid originating from the genome of a living organism. Their genetic material could have been derived from transferable genetic elements such as [[plasmid]]s or [[transposon]]s, which are prone to moving around, exiting, and entering genomes. | *Small viruses with only a few genes may be runaway stretches of nucleic acid originating from the genome of a living organism. Their genetic material could have been derived from transferable genetic elements such as [[plasmid]]s or [[transposon]]s, which are prone to moving around, exiting, and entering genomes. | ||
Revision as of 00:17, 12 December 2006
- This article is about a biological infectious particle; for the computer term, see computer virus. For other uses, see virus (disambiguation).
| Viruses | ||
|---|---|---|
 | ||
| Virus classification | ||
| ||
| Groups | ||
|
I: dsDNA viruses |
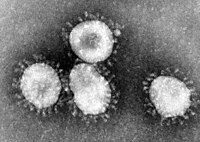
A virus (Latin, poison) is a microscopic particle that can infect the cells of a biological organism. At the most basic level, viruses consist of genetic material contained within a protective protein shell called a capsid; the existence of both genetic material and protein distinguishes them from other virus-like particles such as prions and viroids. They infect a wide variety of organisms: both eukaryotes (such as animals and plants) and prokaryotes (such as bacteria). A virus that infects bacteria is known as a bacteriophage, often shortened to phage. The study of viruses is known as virology. A virologist studies viruses.
Are viruses are living organisms? That question has undergone an extensive argument. Most virologists consider them non-living, as they do not meet all the criteria of the generally accepted definition of life. Like obligate intracellular parasites they lack the means for self-reproduction outside a host cell, but unlike these parasites, viruses are generally not considered to be truly alive. Among other factors, viruses do not possess a cell membrane or metabolise on their own. A definitive answer is still elusive because some organisms considered to be living exhibit characteristics of both living and non-living particles, as viruses do. For those who consider viruses living, viruses are an exception to the cell theory proposed by Theodore Schwann, as viruses are not made up of cells.
Discovery
Viral diseases such as rabies have affected humans for many centuries, but the cause of these diseases was discovered relatively recently. In the early 18th century, the wife of an English ambassador to Turkey observed local women inoculating their children against smallpox. The children subsequently became immune to the disease. In the late 18th century, Edward Jenner observed and studied a milkmaid who had previously caught cowpox was then found to be immune to smallpox, a similar virus.
In the late 19th century Charles Chamberland developed a porcelain filter. This filter was used to study the first documented virus, tobacco mosaic virus. Shortly afterwards, Dimitri Ivanovski published experiments showing that crushed leaf extracts of infected tobacco plants were still infectious even after filtering the bacteria from the solution. At about the same time, several others documented filterable disease-causing agents, with several independent experiments showing that viruses were different from bacteria, yet they could also cause disease in living organisms.
In the early 20th century, Frederick Twort discovered that bacteria itself could be attacked by viruses. Felix d'Herelle, working independently, showed that a preparation of viruses caused areas of cellular death on thin cell cultures spread on agar. Counting the dead areas allowed him to estimate the original number of viruses in the suspension. Finally, in 1935 Wendell Stanley crystallised the tobacco mosaic virus and found it to be mostly protein. A short time later the virus was separated into protein and nucleic acid parts.
Origins
The origins of modern viruses are not entirely clear, and there may not be a single mechanism of origin that can account for all viruses. As viruses do not fossilise well, molecular techniques, rather than the geologic records, have provided the most useful means of hypothesising how they arose. Research in microfossil identification and molecular biology may yet discern fossil evidence dating to the Archean or Proterozoic eons. Two main hypotheses currently exist[1]:
- Small viruses with only a few genes may be runaway stretches of nucleic acid originating from the genome of a living organism. Their genetic material could have been derived from transferable genetic elements such as plasmids or transposons, which are prone to moving around, exiting, and entering genomes.
- Viruses with larger genomes, such as poxviruses, may have once been small cells which parasitised larger host cells. Over time, genes not required by their parasitic lifestyle would have been lost in a streamlining process known as retrograde- or reverse-evolution. Both the bacteria Rickettsia and Chlamydia are living cells which, like viruses, can only reproduce inside host cells. They lend credence to this hypothesis, as they are likely to have lost genes which enabled them to survive outside a host cell in favour of their parasitic lifestyle.
Other infectious particles which are even simpler in structure than viruses include viroids, satellites, and prions.
Classification
Template:Details In taxonomy, the classification of viruses has proved to be rather difficult due to the lack of a fossil record and dispute over whether they are living or non-living. They do not fit easily into any of the domains of biological classification and therefore classification begins at the family rank. However, the domain name of Acytota has been suggested. This would place viruses on a par with the other domains of Eubacteria, Archaea, and Eukarya. Not all families are currently classified into orders, nor all genera classified into families.
As an example of viral classification, the chicken pox virus belongs to family Herpesviridae, subfamily Alphaherpesvirinae and genus Varicellovirus. It remains unranked in terms of order. The general structure is as follows.
The International Committee on Taxonomy of Viruses (ICTV) developed the current classification system and put in place guidelines that put a greater weighting on certain virus properties in order to maintain family uniformity. In determining order, taxonomists should consider the type of nucleic acid present, whether the nucleic acid is single- or double-stranded, and the presence or absence of an envelope. After these three main properties, other characteristics can be considered: the type of host, the capsid shape, immunological properties and the type of disease it causes.
In addition to this classification system, the Nobel Prize-winning biologist David Baltimore devised the Baltimore classification system. This places a virus into one of seven Groups, which distinguish viruses based on their mode of replication and genome type. The ICTV classification system is used in conjunction with the Baltimore classification system in modern virus classification.
Structure
A complete virus particle, known as a virion, is little more than a gene transporter, consisting at the most basic level of nucleic acid surrounded by a protective coat of protein called a capsid. A capsid is composed of proteins encoded by the viral genome and its shape serves as the basis for morphological distinction. Virally coded protein units called protomers will self-assemble to form the capsid, requiring no input from the virus genome - however, a few viruses code for proteins which assist the construction of their capsid. Proteins associated with nucleic acid are more technically known as nucleoproteins, and the association of viral capsid proteins with viral nucleic acid is called a nucleocapsid.
In general, four main morphological virus types can be identified:
| Helical viruses | |
| Helical capsids are composed of a single type of protomer stacked around a central circumference to form an enclosed tube resembling a spiral staircase. This arrangement results in rod-shaped virions which can be short and rigid, or long and flexible. Long helical particles must be flexible in order to prevent forces snapping the structure. The genetic material is housed on the inside of the tube, protected from the outside. Overall, the length of a helical capsid is related to the length of the nucleic acid contained within it, while the diameter is dependent on the overall length and arrangement of protomers. The well-studied tobacco mosaic virus is a helical virus. | |
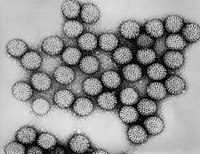
| Icosahedral viruses | |
| Icosahedral capsid symmetry results in a spherical appearance of viruses at low magnification but actually consists of capsomers arranged in a regular geometrical pattern, similar to a soccer ball, hence they are not truly "spherical". Capsomers are ring shaped structures constructed from five to six copies of protomers. These associate via non-covalent bonding to enclose the viral nucleic acid, though generally less intimately than helical capsids, and may involve one type of protomer or more.
Icosahedral architecture was employed by R. Buckminster-Fuller in his geodesic dome, and is the most efficient way of creating an enclosed robust structure from multiple copies of a single protein. The number of proteins required to form a spherical virus capsid is denoted by the T-number[2], where 60×t proteins are necessary. In the case of the hepatitis B virus the T-number is 4, therefore 240 proteins assemble to form the capsid. | |
| Enveloped viruses | |
File:800px-HIV Viron.png Diagram of enveloped HIV |
In addition to a capsid some viruses are able to hijack a modified form of the cell membrane surrounding an infected host cell, thus gaining an outer lipid layer known as a viral envelope. This extra membrane is studded with proteins coded for by the viral genome and host genome, however the lipid membrane itself and any carbohydrates present are entirely host-coded.
The viral envelope can give a virion a few distinct advantages over other "naked" virions, such as protection from enzymes and chemicals. The proteins studded upon it can include glycoproteins functioning as receptor molecules, allowing healthy cells to recognise virions as "friendly" and resulting in the possible uptake of the virion into the cell. Some viruses are so dependent upon their viral envelope that they fail to function if it is removed. |
| Complex viruses | |
| These viruses possess a capsid which is neither purely helical, nor purely icosahedral, and which may possess extra structures such as protein tails or a complex outer wall. Some bacteriophages have a complex structure consisting of an icosahedral head bound to a helical tail, the latter of which may have a hexagonal base plate with many protruding protein tail fibres.
The poxviruses are large, complex viruses which possess unusual morphology. The viral genome is associated with proteins within a central disk structure known as a nucleoid. The nucleoid is surrounded by a membrane and two lateral bodies of unknown function. Covering the virus is an outer envelope with a thick layer of protein studded on its surface. The whole particle is slightly pleiomorphic, ranging from ovoid to brick shape. |
Size
The majority of viruses which have been studied have a capsid diameter between 10 and 300 nanometres. To put viral size into perspective, a medium sized virion next to a flea is roughly equivalent to a human next to a mountain twice the size of Mount Everest. Some filoviruses have a total length of up to 1400 nm, however their capsid diametres are only about 80 nm. While most viruses are unable to be seen with a light microscope, some are larger than the smallest bacteria and can be seen under high magification. Both scanning and transmission electron microscopes are commonly employed to visualise virus particles.
A notable exception to the normal viral size range is the recently discovered mimivirus, with a diameter of 400 nm. They also hold the record for the largest viral genome size, possessing about 1000 genes (some bacteria only possess 400) on a genome approximately 1.2 megabases in length. Their large genome also contains many genes which are conserved in both prokaryotic and eukaryotic genes[3]. The discovery of the virus has led many scientists to reconsider the controversial boundary between living organisms and viruses, which are currently considered as mere mobile genetic elements.
Genetic material
Both DNA and RNA are found in viral species, but generally a species will not contain both. One exception is the human cytomegalovirus, which contains both a DNA core and several mRNA segments. The nucleic acid can be either single- or double-stranded, depending on the species. Therefore viruses as a group contain all four possible types of nucleic acids: double-stranded DNA, single-stranded DNA, double-stranded RNA and single-stranded RNA. Animal virus species have been observed to possess all combinations, whereas plant viruses tend to have single-stranded RNA. Bacteriophages tend to have double-stranded DNA. Also, the nucleic acids can be either linear or a closed loop.
Genome size in terms of the weight of nucleotides varies quite substantially between species. The smallest genomes code for only four proteins and weigh about 106 daltons, while the largest weigh about 108 daltons and code for over one hundred proteins. Some virus species possess abnormal nucleotides, such as hydroxymethylcytosine instead of cytosine, as a normal part of their genome.
For viruses with RNA as their nucleic acid, the strands are said to be either positive-sense (also called plus-strand) or negative-sense (also called minus-strand) depending on whether it is complementary to viral mRNA. Positive-sense viral RNA is identical to viral mRNA and thus can be immediately translated by the host cell. Negative-sense viral RNA is complementary to mRNA and thus must be converted to positive-sense RNA by an RNA polymerase before translation.
All double-stranded RNA genomes and some single-stranded RNA genomes are said to be segmented, or divided into separate parts. Each segment may code for one protein, and they are usually found together in one capsid. Not all segments are required to be in the same virion for the overall virus to be infectious, as can be seen in the brome mosaic virus.
Replication
Viral populations do not grow through cell division, because they are acellular; instead, they use the machinery and metabolism of a host cell to produce multiple copies of themselves. They may have a lytic or a lysogenic cycle, with some viruses capable of carrying out both. A virus can still cause degenerative effects within a cell without causing its death; collectively these are termed cytopathic effects. Released virions can be passed between hosts through either direct contact, often via body fluids, or through a vector. In aqueous environments, viruses float free in the water.
In the lytic cycle, characteristic of virulent phages such as the T4 phage, host cells will be induced by the virus to begin manufacturing the proteins necessary for virus reproduction. As well as proteins, the virus must also direct the replication of new genomes, the technique used for this varies greatly between virus species but depends heavily on the genome type. The final viral product is assembled spontaneously, though it may be aided by molecular chaperones. After the genome has been replicated and the new capsid assembled, the virus causes the cell to be broken open (lysed) to release the virus particles. Some viruses do not lyse the cell but instead exit the cell via the cell membrane in a process known as exocytosis, taking a small portion of the membrane with them as a viral envelope. As soon as the cell is destroyed the viruses have to find a new host.
In contrast, the lysogenic cycle does not result in immediate lysing of the host cell, instead the viral genome integrates into the host DNA and replicates along with it. The virus remains dormant but after the host cell has replicated several times, or if environmental conditions permit it, the virus will become active and enter the lytic phase. The lysogenic cycle allows the host cell to continue to survive and reproduce, and the virus is passed on to all of the cell’s offspring.

Bacteriophages infect specific bacteria by binding to surface receptor molecules and entering the cell. Within a short amount of time, sometimes just minutes, bacterial polymerase starts translating viral mRNA into protein. These proteins go on to become either new virions within the cell, helper proteins which help assembly of new virions, or proteins involved in cell lysis. Viral enzymes aid in the breakdown of the cell membrane, and in the case of the T4 phage, in just over twenty minutes after injection over three hundred phages will be released.
Animal DNA viruses, such as herpesviruses, enter the host via endocytosis, the process by which cells take in material from the external environment. This frequently occurs after chance collision with an appropriate surface receptor on a cell. After penetrating the cell, the viral genome is released from the capsid and host polymerases begin transcribing viral mRNA. New virions are assembled and released either by cell lysis or by budding off the cell membrane.
Animal RNA viruses can be placed into about four different groups depending on their mode of replication. The polarity of the RNA largely determines the replicative mechanism, as well as whether the genetic material is single-stranded or double-stranded. Some RNA viruses are actually DNA based but use an RNA-intermediate to replicate. RNA viruses are heavily dependent upon virally encoded RNA replicase to create copies of their genomes.
A reverse transcribing virus is any virus which replicates using reverse transcription, the formation of DNA from an RNA template. Those viruses containing RNA genomes use a DNA intermediate to replicate, whereas those containing DNA genomes use an RNA intermediate during genome replication. Both types of reverse transcribing viruses use the enzyme reverse transcriptase to carry out the nucleic acid conversion.
Lifeform debate
Argument continues over whether viruses are truly alive. According to the United States Code, they are considered to be micro-organisms in the sense of biological weaponry and malicious use. Scientists however are more divided. They have no trouble classifying a horse as living, but things become complicated as they look at the more simple viruses, viroids and prions. Viruses resemble life in that they possess nucleic acid and can respond to their environment in a limited fashion. They can also reproduce by creating multiple copies of themselves through simple self-assembly.
However, unlike all established types of lifeform, they do not possess a cell structure, regarded as the basic unit of life. Viruses are also absent from the fossil record, making phylogenic relationships difficult to infer. Additionally, although they reproduce, they do not metabolise on their own and therefore require a host cell to replicate and synthesise new products. However, confounding this previous statement is the fact that bacterial species such as Rickettsia and Chlamydia, while living organisms, are also unable to reproduce outside of a host cell.
An argument can be made that all accepted forms of life divide at the cell level via cell division to reproduce, whereas all viruses assemble spontaneously within cells. The comparison is drawn between viral self-assembly and the autonomous growth of non-living crystals. Virus self-assembly within host cells also has implications for the study of the origin of life, as it lends credence to the hypothesis that life could have started as self-assembling organic molecules.
Other questions involve the classification of viruses within the evolutionary tree and its implications – if viruses are considered alive, then the criteria specifying life will have been permanently changed, leading scientists to question what the basic prerequisite of life is. If they are considered living then the prospect of creating artificial life is enhanced, or at least the standards required to call something artificially alive are reduced. If viruses were said to be alive, the question could follow of whether other even smaller infectious particles, such as viroids and prions, would next be considered forms of life.
Viruses and disease
- For more examples of diseases caused by viruses see List of infectious diseases
Examples of common human diseases caused by viruses include the common cold, the flu, chickenpox and cold sores. Serious diseases such as Ebola, AIDS, bird flu and SARS are all also caused by viruses. The relative ability of viruses to cause disease is described in terms of virulence. Other diseases are under investigation as to whether they too have a virus as the causative agent, such as the possible connection between Human Herpesvirus Six (HHV6) and neurological diseases such as multiple sclerosis and chronic fatigue syndrome. Recently it was also shown that cervical cancer is partially caused by papillomavirus, representing evidence in humans of a link existing between cancer and an infective agent[4]. There is current controversy over whether the borna virus, previously thought of as causing neurological disease in horses, could be responsible for psychiatric illness in humans[5].
Viruses have many different mechanisms by which they produce disease in an organism, which largely depends on the species. Mechanisms at the cellular level primarily include cell lysis, the breaking open and subsequent death of the cell. In multicellular organisms, if enough cells die the whole organism will start to suffer the effects. Although many viruses result in the disruption of healthy homeostasis, resulting in disease, they may also exist relatively harmlessly within an organism. An example would include the ability of the herpes simplex virus, which cause coldsores, to remain in a dormant state within the human body.
Epidemics
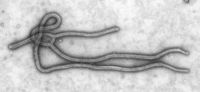
A number of highly lethal viral pathogens are members of the Filoviridae. Filoviruses are filament-like viruses that cause viral hemorrhagic fever, and include the Ebola and Marburg viruses. The Marburg virus attracted widespread press attention in April 2005 for an outbreak in Angola. Beginning in October 2004 and continuing into 2005, the outbreak was the world's worst epidemic of any kind of viral hemorrhagic fever[6].
Native American populations were devastated by contagious diseases, particularly smallpox, brought to the Americas by European colonists. It is unclear how many Native Americans were killed by foreign diseases after the arrival of Columbus in the Americas, but the numbers have been estimated to be close to 70% of the indigenous population[7]. The damage done by this disease may have significantly aided European attempts to displace or conquer the native population. Viruses also cause some of the most dangerous diseases ever known to man, such as smallpox and AIDS.
Detection, purification and diagnosis
In the laboratory, several techniques for growing and detecting viruses exist. Purification of viral particles can be achieved using differential centrifugation, isopycnic centrifugation, precipitation with ammonium sulfate or ethylene glycol, and removal of cell components from a homogenised cell mixture using organic solvents or enzymes to leave the virus particles in solution.
Assays to detect and quantify viruses include:.
- Hemagglutination assays, which quantitatively measure how many virus particles are in a solution of red blood cells by the amount of agglutination the viruses cause between them. This occurs as many viruses are able to bind to the surface of one or more red blood cells.
- Direct counts using an electron microscope. A dilute mixture of virus particles and beads of known size are sprayed onto a special sheet and examined under high magnification. The virions are counted and the number extrapolated to estimate the number of virions in the undiluted mixture.
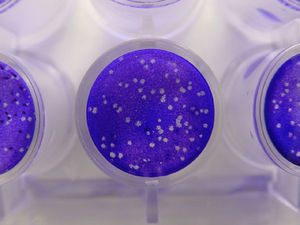
- Plaque assays involve growing a thin layer of host cells onto a culture dish and adding a dilute mixture of virions onto it. The virions will infect and kill the cells they land on, producing holes in the cell layer known as plaques. The number of plaques can be counted and the number of virions estimated from it.
Detection and subsequent isolation of new viruses from patients is a specialised laboratory subject. Normally it requires the use of large facilities, expensive equipment, and trained specialists such as technicians, molecular biologists, and virologists. Often, this effort is undertaken by state and national governments and shared internationally through organizations like the World Health Organization.
Prevention and treatment
Because viruses use the machinery of a host cell to reproduce and also reside within them, they are difficult to eliminate without killing the host cell. The most effective medical approaches to viral diseases so far are vaccinations to provide resistance to infection, and drugs which treat the symptoms of viral infections. Patients often ask for, and physicians often prescribe, antibiotics. These are useless against viruses, and their misuse against viral infections is one of the causes of antibiotic resistance in bacteria. However, in life-threatening situations the prudent course of action is to begin a course of antibiotic treatment while waiting for test results to determine whether the patient's symptoms are caused by a virus or a bacterial infection.
Applications

Life sciences
Viruses are important to the study of molecular and cellular biology as they provide simple systems that can be used to manipulate and investigate the functions of cells. The study and use of viruses have provided valuable information about many aspects of cell biology. For example, viruses have simplified the study of genetics and helped human understanding of the basic mechanisms of molecular genetics, such as DNA replication, transcription, RNA processing, translation, protein transport, and immunology.
Geneticists regularly use viruses as vectors to introduce genes into cells that they are studying. This is useful for making the cell produce a foreign substance, or to study the effect of introducing a new gene into the genome. In similar fashion, virotherapy uses viruses as vectors to treat various diseases, as they can specifically target cells and DNA. It shows promising use in the treatment of cancer and in gene therapy.
Materials science and nanotechnology
In April 2006 scientists at the Massachusetts Institute of Technology (MIT) created nanoscale metallic wires using a genetically-modified virus[8]. The MIT team was able to use the virus to create a working battery with an energy density up to three times more than current materials. The potential exists for this technology to be used in liquid crystals, solar cells, fuel cells, and other electronics in the future.
Weapons
Template:Details The ability of viruses to cause devastating epidemics in human societies has led to the concern that viruses could be weaponized for biological warfare. Further concern was raised by the successful recreation of the infamous 1918 influenza virus in a laboratory[9]. The smallpox virus devastated numerous societies throughout history before its eradication. It currently exists in several secure laboratories in the world, and fears that it may be used as a weapon are not totally unfounded. The modern global human population has almost no established resistance to smallpox; if it were to be released, a massive loss of life could be sustained before the virus was brought under control.
Etymology
The word is from the Latin virus referring to poison and other noxious things, first used in English in 1392. Virulent, from Latin virulentus "poisonous" dates to 1400. A meaning of "agent that causes infectious disease" is first recorded in 1728, before the discovery of viruses by the Russian-Ukrainian biologist Dmitry Ivanovsky in 1892. The adjective viral dates to 1948. Today, virus is used to describe the biological viruses discussed above and also as a metaphor for other parasitically-reproducing things, such as memes or computer viruses (since 1972). The neologism virion or viron is used to refer to a single infective viral particle.
The Latin word is from a Proto-Indo-European root *Template:PIE "to melt away, to flow," used of foul or malodorous fluids. It is a cognate of Sanskrit Template:IAST "poison,", Avestan viš- "poison," Greek ios "poison," Old Church Slavonic višnja "cherry," Old Irish fi "poison," Welsh gwy "fluid"; Latin viscum (see viscous) "sticky substance" is also from the same root.
The English plural form of virus is viruses. No reputable dictionary gives any other form, including such "reconstructed" Latin plural forms as viri (which actually means men), and no plural form appears in the Latin corpus (See plural of virus). The word does not have a traditional Latin plural because its original sense, poison is a mass noun like the English word furniture, and, as pointed out above, English use of virus to denote the agent of a disease predates the discovery that these agents are microscopic parasites and thus in principle countable.
References
Citations
- ↑ Prescott, L. (1993). Microbiology, Wm. C. Brown Publishers, ISBN 0-697-01372-3
- ↑ http://rhino.bocklabs.wisc.edu/cgi-bin/virusworld/htdocs.pl?docname=triangulation.html
- ↑ http://www.stanford.edu/group/virus/mimi/2005/Genome.htm
- ↑ http://news.bbc.co.uk/2/hi/health/medical_notes/429762.stm
- ↑ http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=PubMed&cmd=Retrieve&list_uids=10089006&dopt=Abstract
- ↑ http://news.bbc.co.uk/2/hi/africa/4397891.stm
- ↑ http://www.historylink.org/essays/output.cfm?file_id=5100
- ↑ http://web.mit.edu/newsoffice/2006/virus-battery.html
- ↑ http://www.cdc.gov/OD/OC/MEDIA/pressrel/r051005.htm
Further reading
- Villarreal, L. P. (2005) Viruses and the Evolution of Life. ASM Press, Washington DC ISBN 1-55581-309-7
- Icosahedral virus structure
- All the Virology on the WWW
- University of Leicester online notes - Virus Structure
- Chronic Active Human Herpesvirus-6 (HHV-6) Infection: A New Disease Paradigm
- Gelderblom, Hans R. (1996). 41. Structure and Classification of Viruses in Medical Microbiology 4th ed. Samuel Baron ed. The University of Texas Medical Branch at Galveston. ISBN 0-9631172-1-1
- Radetsky, Peter (1994). The Invisible Invaders: Viruses and the Scientists Who Pursue Them. Backbay Books, ISBNs 0316732168 (hc), 0316732176 (pb).
- Theiler, Max and Downs, W. G. (1973). The Arthropod-Borne Viruses of Vertebrates: An Account of the Rockefeller Foundation Virus Program 1951-1970. Yale University Press.
- Template:NCBI-scienceprimer
See also
Template:Wikibookspar Template:Commons Template:Wiktionary
External links
- Chart of viral pathogens which contribute to indoor air pollution
- Viruses: The new cancer hunters - An IsraCast article on virotherapy
- The Big Picture Book of Viruses - Pictures and general information on many viruses
- Scientific American Magazine (October 2003 Issue) Tumor-Busting Viruses
- Detailed genomic and bioinformatic information about Category A, B, and C priority pathogens at NIH-funded database.
- Assorted information about Viruses.
ar:فيروس bg:Вирус ca:Virus cs:Virus de:Virus et:Viirused es:Virus eo:Viruso eu:Birus fa:ویروس fr:Virus ko:바이러스 hi:वायरस id:Virus it:Virus (biologia) he:נגיף la:Virus biologicum lv:Vīruss lt:Virusas hu:Vírus (biológia) mk:Вирус mr:विषाणू ms:Virus nl:Virus (biologie) ja:ウイルス no:Virus nn:Virus pl:Wirus (biologia) pt:Vírus (biologia) ro:Virus ru:Вирусы simple:Virus sk:Vírus sl:Virus sr:Вирус su:Virus fi:Virukset sv:Virus ta:அதி நுண் நச்சுயிர் th:ไวรัส vi:Virus tr:Virüs (Biyoloji) ur:وائرس yi:וויירוס zh:病毒