Arabidopsis thaliana (Thale Cress): Difference between revisions
imported>Chris Day (New page: {{taxobox |name = ''Arabidopsis thaliana'' |image = Arabidopsis thaliana.jpg |regnum = Plantae |unranked_divisio = Angiosperms |unranked_classis = Eudicots |unranked_ordo = [[R...) |
imported>Chris Day m (Arabidopsis thaliana moved to Arabidopsis thaliana (Thale Cress)) |
(No difference)
| |
Revision as of 08:11, 24 September 2008
| Arabidopsis thaliana | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
 | ||||||||||||||
| Scientific classification | ||||||||||||||
| ||||||||||||||
| Binomial name | ||||||||||||||
| Arabidopsis thaliana (L.) Heynh. | ||||||||||||||
 | ||||||||||||||
| Synonyms | ||||||||||||||
|
Arabis thaliana |
Arabidopsis thaliana (A-ra-bi-dóp-sis tha-li-á-na; thale cress, mouse-ear cress or Arabidopsis), is a small flowering plant native to Europe, Asia, and northwestern Africa.[1] A spring annual with a relatively short life cycle, Arabidopsis is popular as a model organism in plant biology and genetics. Its genome is one of the smallest plant genomes and was the first plant genome to be sequenced. Arabidopsis is a popular tool for understanding the molecular biology of many plant traits, including flower development and light sensing.
Habitat, morphology, and life cycle
Arabidopsis is native to Europe, Asia, and northwestern Africa. It is an annual (rarely biennial) plant usually growing to 20–25 cm tall. The leaves form a rosette at the base of the plant, with a few leaves also on the flowering stem. The basal leaves are green to slightly purplish in colour, 1.5–5 cm long and 2–10 mm broad, with an entire to coarsely serrated margin; the stem leaves are smaller, unstalked, usually with an entire margin. Leaves are covered with small unicellular hairs (called trichomes). The flowers are 3 mm in diameter, arranged in a corymb; their structure is that of the typical Brassicacaea. The fruit is a siliqua 5–20 mm long, containing 20–30 seeds.[2][3][4][5] Roots are simple in structure, with a single primary root that grows vertically downwards, later producing smaller lateral roots. These roots form interactions with rhizosphere bacteria such as Bacillus megaterium.[6]
Arabidopsis can complete its entire life cycle in six weeks. The central stem that produces flowers grows after about three weeks, and the flowers naturally self-pollinate. In the lab Arabidopsis may be grown in petri plates or pots, under fluorescent lights or in a greenhouse.[7]
Use as a model organism
Arabidopsis is widely used as one of the model organisms for studying plant sciences, including genetics and plant development.[8][9] It plays the role for agricultural sciences that mice and fruit flies (Drosophila) play in animal biology. Although Arabidopsis thaliana has little direct significance for agriculture, it has several traits that make it a useful model for understanding the genetic, cellular, and molecular biology of flowering plants.
The small size of its genome make Arabidopsis thaliana useful for genetic mapping and sequencing — with about 157 million base pairs[10] and five chromosomes, Arabidopsis has one of the smallest genomes among plants. It was the first plant genome to be sequenced, completed in 2000 by the Arabidopsis Genome Initiative.[11] Much work has been done to assign functions to its 27,000 genes and the 35,000 proteins they encode.[12]
The plant's small size and rapid life cycle are also advantageous for research. Having specialized as a spring ephemeral, it has been used to found several laboratory strains that take about six weeks from germination to mature seed. The small size of the plant is convenient for cultivation in a small space and it produces many seeds. Further, the selfing nature of this plant assists genetic experiments. Also, as an individual plant can produce several thousand seeds, each of the above criteria leads to Arabidopsis thaliana being valued as a genetic model organism.
Finally, plant transformation in Arabidopsis is routine, using Agrobacterium tumefaciens to transfer DNA to the plant genome. The current protocol, termed "floral-dip", involves simply dipping a flower into a solution containing Agrobacterium, the DNA of interest, and a detergent.[13] This method avoids the need for tissue culture or plant regeneration.
History of Arabidopsis research
The first mutant in Arabidopsis was documented in 1873 by Alexander Braun, describing a double flower phenotype (the mutated gene was likely Agamous, cloned and characterized in 1990).[14] However, it was not until 1943 that Friedrich Laibach (who had published the chromosome number in 1907) proposed Arabidopsis as a model organism.[15] His student Erna Reinholz published her thesis on Arabidopsis in 1945, describing the first collection of Arabidopsis mutants that they generated using x-ray mutagenesis. Laibach continued his important contributions to Arabidopsis research by collecting a large number of ecotypes. With the help of Albert Kranz, these were organised into the current ecotype collection of 750 natural accessions of Arabidopsis thaliana from around the world.
In the 1950s and 1960s John Langridge and George Rédei played an important role in establishing arabidopsis as a useful organism for biological laboratory experiments. Rédei wrote several scholarly reviews instrumental in introducing the model to the scientific community. The start of the arabidopsis research community dates to a newsletter called Arabidopsis Information Service (AIS), established in 1964. The first International Arabidopsis Conference was held in 1965, in Göttingen, Germany.
In the 1980s Arabidopsis started to become widely used in plant research laboratories around the world. It was one of several candidates that included maize, petunia and tobacco.[15] The latter two were attractive since they were easily transformable with the then current technologies, while maize was a well established genetic model for plant biology. The breakthrough year for Arabidopsis as the preferred model plant came in 1986 when T-DNA mediated transformation was first published and this coincided with the first gene to be cloned and published.[16][17]
Research
Flower development
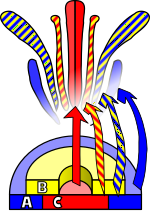
Arabidopsis has been extensively studied as a model for flower development. The developing flower has four basic organs: sepals, petals, stamens, and carpels (which go on to form pistils). These organs are arranged in a series of whorls: four sepals on the outer whorl, followed by six petals inside this, six stamens, and a central carpel region. Homeotic mutations in Arabidopsis result in the change of one organ to another — in the case of the Agamous mutation, for example, stamens become petals and carpels are replaced with a new flower, resulting in a recursively repeated sepal-petal-petal pattern.
Observations of homeotic mutations led to the formulation of the ABC model of flower development by E. Coen and E. Meyerowitz.[18] According to this model floral organ identity genes are divided into three classes: class A genes (which affect sepals and petals), class B genes (which affect petals and stamens), and class C genes (which affect stamens and carpels). These genes code for transcription factors that combine to cause tissue specification in their respective regions during development. Although developed through study of Arabidopsis flowers, this model is generally applicable to other flowering plants.
Light sensing
The photoreceptors phytochrome A, B, C, D and E mediate red light based phototropic response. Understanding the function of these receptors has helped plant biologists understand the signalling cascades that regulate photoperiodism, germination, de-etiolation and shade avoidance in plants.
Arabidopsis was used extensively in the study of the genetic basis of phototropism, chloroplast alignment, and stomatal aperture and other blue light-influenced processes.[19] These traits respond to blue light, which is perceived by the phototropin light receptors. Arabidopsis has also been important in understanding the functions of another blue light receptor, cryptochrome, which is especially important for light entrainment to control the plants circadian rhythms.[20]
Light response was even found in roots, which were thought not to be particularly sensitive to light. While gravitropic response of Arabidopsis root organs is their predominant tropic response, specimens treated with mutagens and selected for the absence of gravitropic action showed negative phototropic response to blue or white light, and positive response to red light.[21]
Non-Mendelian inheritance
In 2005, scientists at Purdue University proposed that Arabidopsis possessed an alternative to previously known mechanisms of DNA repair, which one scientist called a "parallel path of inheritance". It was observed in mutations of the HOTHEAD gene. Plants mutant in this gene exhibit organ fusion, and pollen can germinate on all plant surfaces, not just the stigma. After spending over a year eliminating simpler explanations, it was indicated that the plants "cached" versions of their ancestors' genes going back at least four generations, and used these records as templates to correct the HOTHEAD mutation and other single nucleotide polymorphisms. The initial hypothesis proposed that the record may be RNA-based[22] Since then, alternative models have been proposed which would explain the phenotype without requiring a new model of inheritance[23][24] More recently the whole phenomenon is being challenged as a being a simple artifact of pollen contamination.[25] "When Jacobsen took great pains to isolate the plants, he couldn't reproduce the [reversion] phenomenon", notes Steven Henikoff.[26] In response to the new finding, Lolle and Pruitt agree that Peng et al. did observe cross-pollination but note that some of their own data, such as double reversions of both mutant genes to the regular form, cannot be explained by cross pollination.[27]
Multigen
This is an ongoing experiment on the International Space Station, it is being performed by the European Space Agency. The goals are to study the growth and reproduction of plants from seed to seed in microgravity.
See also
References
- ↑ Germplasm Resources Information Network: Arabidopsis thaliana
- ↑ Flora of NW Europe: Arabidopsis thaliana
- ↑ Blamey, M. & Grey-Wilson, C. (1989). Flora of Britain and Northern Europe. ISBN 0-340-40170-2
- ↑ Flora of Pakistan: Arabidopsis thaliana
- ↑ Flora of China: Arabidopsis thaliana
- ↑ López-Bucio J, Campos-Cuevas JC, Hernández-Calderón E, et al (2007). "Bacillus megaterium rhizobacteria promote growth and alter root-system architecture through an auxin- and ethylene-independent signaling mechanism in Arabidopsis thaliana". Mol. Plant Microbe Interact. 20 (2): 207–17. DOI:10.1094/MPMI-20-2-0207. PMID 17313171. Research Blogging.
- ↑ D.W. Meinke, J.M. Cherry, C. Dean, S.D. Rounsley, M. Koornneef (1998). "Arabidopsis thaliana: A Model Plant for Genome Analysis". Science 282 (5389): 662–682. DOI:10.1126/science.282.5389.662. Research Blogging.
- ↑ Rensink WA, Buell CR (2004). "Arabidopsis to rice. Applying knowledge from a weed to enhance our understanding of a crop species". Plant Physiol. 135 (2): 622–9. DOI:10.1104/pp.104.040170. PMID 15208410. Research Blogging.
- ↑ Coelho SM, Peters AF, Charrier B, et al (2007). "Complex life cycles of multicellular eukaryotes: new approaches based on the use of model organisms". Gene 406 (1-2): 152–70. DOI:10.1016/j.gene.2007.07.025. PMID 17870254. Research Blogging.
- ↑ Bennett, M. D., Leitch, I. J., Price, H. J., & Johnston, J. S. (2003). "Comparisons with Caenorhabditis (100 Mb) and Drosophila (175 Mb) Using Flow Cytometry Show Genome Size in Arabidopsis to be 157 Mb and thus 25% Larger than the Arabidopsis Genome Initiative Estimate of 125 Mb". Annals of Botany 91: 547–557. DOI:10.1093/aob/mcg057. PMID 12646499. Research Blogging.
- ↑ The Arabidopsis Genome Initiative (2000). "Analysis of the genome sequence of the flowering plant Arabidopsis thaliana". Nature 408: 796–815. DOI:10.1038/35048692. PMID 11130711. Research Blogging.
- ↑ Integr8 - A.thaliana Genome Statistics:.
- ↑ Zhang X, Henriques R, Lin SS, Niu QW, Chua NH (2006). "Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method". Nat Protoc 1 (2): 641–6. DOI:10.1038/nprot.2006.97. PMID 17406292. Research Blogging.
- ↑ M.F. Yanofsky, H. Ma, J.L. Bowman, G.N. Drews, K.A. Feldmann & E.M. Meyerowitz (1990). "The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors". Nature 346: 35–39. DOI:10.1038/346035a0. PMID 1973265. Research Blogging.
- ↑ 15.0 15.1 E.M. Meyerowitz (2001). "Prehistory and History of Arabidopsis Research". Plant Physiology 125: 15–19. DOI:10.1038/346035a0. PMID 11154286. Research Blogging.
- ↑ Lloyd AM, Barnason AR, Rogers SG, Byrne MC, Fraley RT, Horsch RB (1986). "Transformation of Arabidopsis thaliana with Agrobacterium tumefaciens". Science 234: 464–466. DOI:10.1126/science.234.4775.464. PMID 17792019. Research Blogging.
- ↑ Chang C, Meyerowitz EM (1986). "Molecular cloning and DNA sequence of the Arabidopsis thaliana alcohol dehydrogenase gene". Proc Natl Acad Sci USA 83: 1408–1412. DOI:10.1073/pnas.83.5.1408. PMID 2937058. Research Blogging.
- ↑ Coen, Henrico S.; Elliot M. Meyerowitz (1991). "The war of the whorls: Genetic interactions controlling flower development". Nature 353: 31–37. DOI:10.1038/353031a0. PMID 1715520. Research Blogging.
- ↑ Sullivan JA, Deng XW (2003). "From seed to seed: the role of photoreceptors in Arabidopsis development". Dev. Biol. 260 (2): 289–97. DOI:10.1016/S0012-1606(03)00212-4. PMID 12921732. Research Blogging.
- ↑ Más P (2005). "Circadian clock signaling in Arabidopsis thaliana: from gene expression to physiology and development". Int. J. Dev. Biol. 49 (5-6): 491–500. DOI:10.1387/ijdb.041968pm. PMID 16096959. Research Blogging.
- ↑ Ruppel NJ, Hangarter RP, Kiss JZ (2001). "Red-light-induced positive phototropism in Arabidopsis roots". Planta 212 (3): 424–30. DOI:10.1007/s004250000410. PMID 11289607. Research Blogging.
- ↑ Lolle SJ, Victor JL, Young JM, Pruitt RE (2005). "Genome-wide non-mendelian inheritance of extra-genomic information in Arabidopsis". Nature 434: 505–9. DOI:10.1038/nature03380. PMID 15785770. Research Blogging. Washington Post summary.
- ↑ Chaudhury, A. (2005). "Hothead healer and extragenomic information". Nature 437: E1–E2. DOI:10.1038/nature04062. PMID 16136082. Research Blogging.
- ↑ Comai L, Cartwright RA (2005). "A toxic mutator and selection alternative to the non-Mendelian RNA cache hypothesis for hothead reversion". Plant Cell 17: 2856–8. DOI:10.1105/tpc.105.036293. PMID 16267378. Research Blogging. summary
- ↑ Peng P., et al. (2006). "Plant genetics: Increased outcrossing in hothead mutants". Nature 443: E8–E9. DOI:10.1038/nature05251. PMID 17006468. Research Blogging.
- ↑ Pennisi E (2006). "Genetics. Pollen contamination may explain controversial inheritance". Science 313: 1864. DOI:10.1126/science.313.5795.1864. PMID 17008492. Research Blogging.
- ↑ Lolle S. J., et al. (2006). "Increased outcrossing in hothead mutants (Reply)". Nature 443: E8–E9. DOI:10.1038/nature05252. PMID 17006468. Research Blogging.
External links
- The European Arabidopsis Stock Centre
- The Arabidopsis Information Resource (TAIR)
- The Arabidopsis Book - comprehensive electronic book
- Arabidopsis thaliana: another "model organism" from Kimball's Biology Pages
- Salk Institute Genomic Analysis Laboratory
- Danish biotech firm Aresa Biotection uses a genetically modified thale cress to detect compounds like explosives (land mines)
- What Makes Plants Grow? The Arabidopsis genome knows Featured article in Genome News Network
- Thale cress goes on the defensive Netherlands Organisation for Scientific Research
- Multigen at NASA.gov