Lactococcus lactis: Difference between revisions
imported>Julie Evans No edit summary |
imported>Julie Evans No edit summary |
||
| Line 22: | Line 22: | ||
L. lactis is a circular chromosome, its genome contains 2,365,589 base pairs. Protein-coding genes represent 86% of the genome, stable RNA 1.4%, and noncoding regions 12.6%. These percentages are very similar to other bacteria genome. Biological functions have been assigned to 64% of the genome, 20.1% have similar genome to other unknown proteins, however, 15% of the genome is unidentifiable. This 15% is hypothesized to contain the traits specific to L.lactis. | L. lactis is a circular chromosome, its genome contains 2,365,589 base pairs. Protein-coding genes represent 86% of the genome, stable RNA 1.4%, and noncoding regions 12.6%. These percentages are very similar to other bacteria genome. Biological functions have been assigned to 64% of the genome, 20.1% have similar genome to other unknown proteins, however, 15% of the genome is unidentifiable. This 15% is hypothesized to contain the traits specific to L.lactis. | ||
==Cell structure and metabolism== | |||
"Lactococcus lactis" can function in both aerobic and anaerobic environments. The bacterium's primitive source of energy is produced anaerobically, which results in the accumulation of lactic acid. The deprivation of oxygen leads the glycolysis process to breakdown carbohydrates into pyruvate which then converts into lactic acid. This process is only possible through the production of the lactate dehydrogenase enzyme and NAD. Lactate is transported to the median which causes the efflux of protons, resulting in the appropriate membrane potential for energy production. Some strains of L. lactis are capable of growing under aerobic conditions, when oxygen and a heme source is present new traits are observed, such as increased growth index, resistance to oxidative and acid stress, and long-termed endurance at low temperatures. Along with heme source, the presence of NADH-oxidases has been related to the presence of NADH-oxidases. | |||
Revision as of 16:03, 1 May 2009
| Lactococcus lactis | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| [[image:|200px|]] | ||||||||||||||
| Scientific classification | ||||||||||||||
| ||||||||||||||
| Binomial name | ||||||||||||||
| Lactococcus lactis |
Description and significance
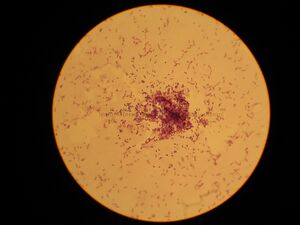
"Lactococcus Lactis" is a non pathogenic,Gram-positive bacteria known as one of the most important microbes in dairy food production. Lactococci lactis, which is closely related to the strptococcus genus, has two subspecies,L. lactis subsp. and Lactococcus cremoris subsp. They are used to make soft and hard cheese respectively. L.Lactis group in pairs or short chains with a length of about 1.2µm by 1.5µm. Lactococci are immotile, nonsporelating, and are usually spherical or ovoid cells. L. Lactis is the starter culture for the production of fermented dairy products such as milk, cheese, yogurt, etc.Lactococcus is considered a opurtunistic pathogen, because of its ability to convert almost all it's sugar into lactic acid. Lactic acid produced by the the microbe curdles the milk which seperates into curds inturn are used to produce cheese and whey.In nature, L. lactis is found to be inactive on plant surfaces and on to multiply in the gastrointestinal tract after being swallowed by an animal.
Genome structure
L. lactis' genome has been sequenced to investigate, what specificly, is its essential role in dairy fermentation. Unexpected features of the analyzed genome have been disclosed, genes are seen to enable the bacterium to perform aerobic respiration along with horizontal gene transfer by transformation. Decades of research has been attributed to the genome sequencing and comparative genomics of L. lactis to unveil the characteristics of L.lactis that can specifically develop flavor, and improve quality and preservation of dairy products such as cheese.
L. lactis is a circular chromosome, its genome contains 2,365,589 base pairs. Protein-coding genes represent 86% of the genome, stable RNA 1.4%, and noncoding regions 12.6%. These percentages are very similar to other bacteria genome. Biological functions have been assigned to 64% of the genome, 20.1% have similar genome to other unknown proteins, however, 15% of the genome is unidentifiable. This 15% is hypothesized to contain the traits specific to L.lactis.
Cell structure and metabolism
"Lactococcus lactis" can function in both aerobic and anaerobic environments. The bacterium's primitive source of energy is produced anaerobically, which results in the accumulation of lactic acid. The deprivation of oxygen leads the glycolysis process to breakdown carbohydrates into pyruvate which then converts into lactic acid. This process is only possible through the production of the lactate dehydrogenase enzyme and NAD. Lactate is transported to the median which causes the efflux of protons, resulting in the appropriate membrane potential for energy production. Some strains of L. lactis are capable of growing under aerobic conditions, when oxygen and a heme source is present new traits are observed, such as increased growth index, resistance to oxidative and acid stress, and long-termed endurance at low temperatures. Along with heme source, the presence of NADH-oxidases has been related to the presence of NADH-oxidases.