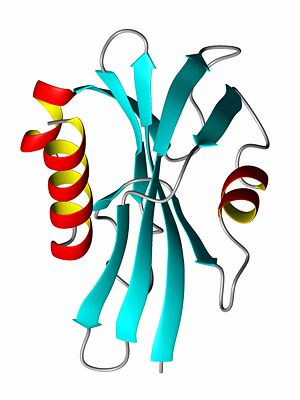
Protein structure
Proteins participate in numerous functions within living organisms, and protein function is largely dependent on the protein structure. Many diseases are caused by proteins whose structures have been modified, either due to amino acid substitutions (sicle cell anemia, cancer) resulting from DNA changes or by incorrect protein folding (alzheimer's).
Basis of protein structure
A Protein is a linear polymer of different amino acids, whose properties vary. To a large degree the shape of a protein is determined by the hydrophobic (water hating) amino acids which prefer not to be solvated and by the hydrophilic (water loving) amino acids, which like to be exposed to water. Hydrogen bonding also plays a significant role, especially in regards to secondary structure.
Types of secondary structure
Some proteins, usually those with a very high percentage of hydrophilic amino acids, are natively unfolded, or are said to have a random coil structure. Many hormone receptor proteins, or large portions of them, are natively unfolded. Most of the proteins for which structures has been determined have secondary structures consisting largely of helices or beta-sheets. A helix is generally a 310-helix (rarely) or an alpha-helix, depending on the degree of twist in the protein backbone. By forming a helix, a protein forms energetically favorable hydrogen bonds between a carboxyl oxygen atom at amino acid position i with the amide proton atom at position i+4 (see figure). Often, one side of the helix is hydrophobic while the other is hydrophilic. The other major element of secondary structure is the beta-sheet which is comprised of individual beta-strands. In a beta-strand, the protein backbone atoms (N,Ca and C) form a zig-zag pattern (see figure). When two beta-strands interact, favorable hydrogen bonds are formed to stabilize the structure. The ends of beta-strands often end with beta-turns (read below).
Tertiary structure
A protein's tertiary structure describes how all of the secondary structure elements fold upon one another. In order to bury the hydrophobic amino acid side chains (or residual groups), the secondary structural elements collapse upon each other so that most of the hydrophobic side chains are secluded away from the solvent, while the hydrophlic amino acids are exposed to the solvent. Thus, a beta-sheet might be protected on both sides by alpha-helices such that the polar side of the helix is exposed to the solvent while the non-polar, hydrophic side of the helix interacts with the hydrophobic residues of the beta-sheet.
Quaternary structure
Quarternary structure describes the interaction (structure) of a group of proteins. Many proteins exist in dimer form, meaning that two molecules of protein bind to each other. Many DNA binding proteins are homodimers. Other proteins form homotrimers and homotetramers. If different protein bind together, they are referred to as hetero-dimers, hetero-trimers, and so forth. Many biological processes occur when dozens of proteins or proteins/DNA/RNA molecules bind together to form a quaternary structure.