Vibrio harveyi: Difference between revisions
imported>John J. Dennehy No edit summary |
imported>John J. Dennehy |
||
| Line 16: | Line 16: | ||
==Description and Significance== | ==Description and Significance== | ||
Vibrio harveyi is found in marine environments (mainly in tropical locations). This bacteria may be either free-living or in symbiosis with marine life like their close relations, Vibrio fischeri. V. harveyi’s 16s RNA classifies them in the Proteobacteria phylum, and their bioluminescence places them in the Vibrionaceae family.<ref name=one>Classification. (2007). Vibrio harveyi. Retrieved April 10, 2009, from Microbe Wiki Web site:http://microbewiki.kenyon.edu/index.php/Vibrio_harveyi</ref>, <ref name=eight>Vibrio harveyi genome [Data file]. (n.d.). Retrieved April 18, 2009, from Kegg Web site:http://www.genome.jp/kegg-bin/show_organism?org=vha</ref> Non-sporulating rods (0.5 microns x 2 microns) help to maintain their structure, while polar flagella whose sheath is an extension of their outer membrane help the bacteria to move. V. harveyi produces an enzyme (luciferase) that generates light seen in their characteristic bioluminescent capabilities, whose functioning is dependent on the cell concentration, and works via quorum sensing. The use of quorum sensing and bioluminescence by V. harveyi is closely studied because of its aid in understanding how bacteria communicate with their environment, detect multiple environmental cues, and use this information as a means of regulating gene expression.<ref name=two>Background. (n.d.). Vibrio harveyi. Retrieved April 10, 2009, from Washington University in St. Louis School of Medicine Genome Sequencing Center Web site: http://genome.wustl.edu/genome.cgl?GENOME=Vibrio%20harveyi</ref> Furthermore, V. harveyi causes some diseases which affect agriculture and cause economic losses; understanding their functioning may help to alleviate these problems as well.<ref name=six>Rao, A. V. (2008, September 12). Vibriosis in shrimp aquaculture. Retrieved April 10, 2009, from http://www.engormix/e_articles_view.asp?art=1179&AREA=ACU-165</ref> Also, the potential for the development of anti-microbial drugs can be investigated with V. harveyi.<ref name=three>Bassler, B. (n.d.). Cell-to-cell communication in bacteria. Retrieved April 10, 2009, from Department of Molecular Biology Princeton University Web site:http://www.molbio.princeton.edu/index.php?option=content&task=view&id=27</ref> | Vibrio harveyi is found in marine environments (mainly in tropical locations). This bacteria may be either free-living or in symbiosis with marine life like their close relations, Vibrio fischeri. V. harveyi’s 16s RNA classifies them in the Proteobacteria phylum, and their bioluminescence places them in the Vibrionaceae family.<ref name=one>Classification. (2007). Vibrio harveyi. Retrieved April 10, 2009, from Microbe Wiki Web site:http://microbewiki.kenyon.edu/index.php/Vibrio_harveyi</ref>, <ref name=eight>Vibrio harveyi genome [Data file]. (n.d.). Retrieved April 18, 2009, from Kegg Web site:http://www.genome.jp/kegg-bin/show_organism?org=vha</ref> Non-sporulating rods (0.5 microns x 2 microns) help to maintain their structure, while polar flagella whose sheath is an extension of their outer membrane help the bacteria to move. V. harveyi produces an enzyme (luciferase) that generates light seen in their characteristic bioluminescent capabilities, whose functioning is dependent on the cell concentration, and works via quorum sensing. The use of quorum sensing and bioluminescence by V. harveyi is closely studied because of its aid in understanding how bacteria communicate with their environment, detect multiple environmental cues, and use this information as a means of regulating gene expression.<ref name=two>Background. (n.d.). Vibrio harveyi. Retrieved April 10, 2009, from Washington University in St. Louis School of Medicine Genome Sequencing Center Web site: http://genome.wustl.edu/genome.cgl?GENOME=Vibrio%20harveyi</ref> Furthermore, V. harveyi causes some diseases which affect agriculture and cause economic losses; understanding their functioning may help to alleviate these problems as well.<ref name=six>Rao, A. V. (2008, September 12). Vibriosis in shrimp aquaculture. Retrieved April 10, 2009, from http://www.engormix/e_articles_view.asp?art=1179&AREA=ACU-165</ref> Also, the potential for the development of anti-microbial drugs can be investigated with V. harveyi.<ref name=three>Bassler, B. (n.d.). Cell-to-cell communication in bacteria. Retrieved April 10, 2009, from Department of Molecular Biology Princeton University Web site:http://www.molbio.princeton.edu/index.php?option=content&task=view&id=27</ref> | ||
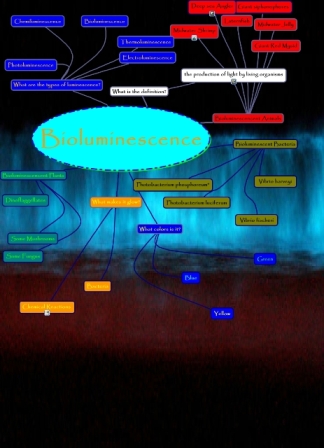
[[Image:Bioluminescence_chart.jpg|center|This tree shows the many areas of life in which bioluminescence exists, the different ways in which it is seen, and the amount of variation there is in a topic that has such strict requirements.]] | |||
==Genome Structure== | ==Genome Structure== | ||
Revision as of 11:58, 18 May 2009
| Vibrio harveyi | ||||||
|---|---|---|---|---|---|---|
 | ||||||
| Scientific classification | ||||||
| ||||||
| Binomial name | ||||||
| Vibrio harveyi |
Description and Significance
Vibrio harveyi is found in marine environments (mainly in tropical locations). This bacteria may be either free-living or in symbiosis with marine life like their close relations, Vibrio fischeri. V. harveyi’s 16s RNA classifies them in the Proteobacteria phylum, and their bioluminescence places them in the Vibrionaceae family.[1], [2] Non-sporulating rods (0.5 microns x 2 microns) help to maintain their structure, while polar flagella whose sheath is an extension of their outer membrane help the bacteria to move. V. harveyi produces an enzyme (luciferase) that generates light seen in their characteristic bioluminescent capabilities, whose functioning is dependent on the cell concentration, and works via quorum sensing. The use of quorum sensing and bioluminescence by V. harveyi is closely studied because of its aid in understanding how bacteria communicate with their environment, detect multiple environmental cues, and use this information as a means of regulating gene expression.[3] Furthermore, V. harveyi causes some diseases which affect agriculture and cause economic losses; understanding their functioning may help to alleviate these problems as well.[4] Also, the potential for the development of anti-microbial drugs can be investigated with V. harveyi.[5]
Genome Structure
The Vibrio harveyi genome has been sequenced up to 8x magnification. Through sequencing it has been determined that the Vibrio harveyi genome consists of two chromosomes and one plasmid. Chromosome I contains 3,765,351 nucleotides, chromosome II contains 2,204,018 nucleotides, and the plasmid contains 89,008 nucleotides.[3] Chromosomes I, II, and the plasmid are all circular. The entire genome contains 6040 genes that encode proteins and 166 genes that encode RNA.[2]
Cell Structure and Metabolism
Vibrio harveyi is a gram negative, facultative anaerobic bacteria with non-sporulating rods and polar flagella. As a gram negative bacteria, it has an thin inner layer of peptidoglycan, surrounded by a periplasmic space, which is surrounded by a thick peptidoglycan layer called the outer membrane and is more permeable than the plasma membrane. The outer membrane is composed of lipids, lipoproteins, and lipopolysaccharides. The polar flagella of the bacteria help it to move in a run and tumble motion while employing chemotaxis (moving towards chemical attractants and away from chemical repellants). As a facultative anaerobe, these bacteria will thrive most in environments with oxygen, but are able to survive in environments lacking oxygen as well.[3]
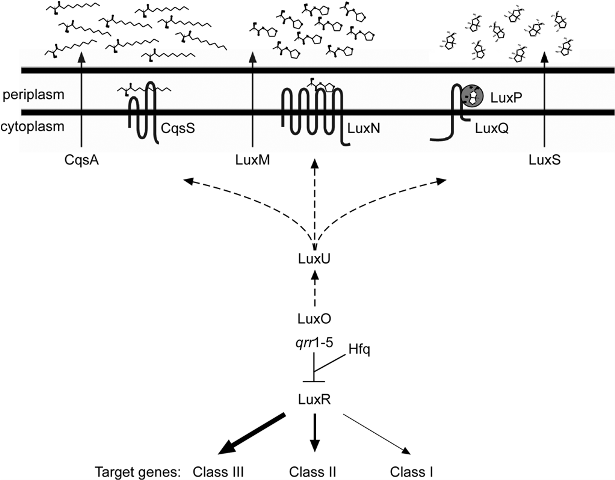
The most distinctive characteristic of V. harveyi (and all bacteria of the vibrionaceae family) is their ability for bioluminescence. This occurs through quorum sensing—a way for the cells to communicate with each other. The bioluminescence is dependent on the concentration of the cells because the necessary enzyme (luciferase) will only be produced in high enough cell concentrations via a series of chemical reactions induced by autoinducers (AI).[3] In addition to bioluminescence, this bacteria also controls some virulence factors, sporulation, and conjugation. Regardless of the purpose of this mechanism, it allows the bacteria to function as a multicellular organism by one of two pathways; an A-1 circuit establishes intra-species communication while an A-2 circuit establishes inter-species communication.[5] When cell density is low, phophorylated LuxO (a response-regulator protein) activates the transcription of genes that encode RNAs that ultimately destabilize and prevent the transcription of LuxR (a quorum-sensing regulatory protein). When the autoinducer levels are raised the phospho relay pathway is reversed, thus decreasing LuxO, increasing LuxR, and allowing for the expression of genes involved in quorum-sensing.[6] In addition to autoinducers, a protein termed Hfq was found in Vibrio harveyi (and V. cholera). Hfq is responsible for mediating interactions between sRNAs and certain mRNAs with the purpose of altering the stability of the target molecules to control gene expression, and also to control the stability of the particular mRNA that codes for LuxR, again to control the expression of quorum sensing genes.[7] Quorum sensing is an activity that is only possible in the presence of many cells, and is ineffective in one or few cells.[5]
Ecology
Aquaculture is the fastest-growing food industry world-wide, and Vibrio harveyi is responsible for many of the diseases that affect this culture (and ultimately cause many economic losses). One such disease is vibriosis, which is responsible for the sickness and death of many captivated shrimp. This disease occurs when bacteria (like V. harveyi) which are tolerated at low levels in shrimp are multiplied as a result of an environmental trigger, or when larger than normal amounts of bacteria penetrate the shrimp’s protective barriers. Though this disease is seen predominantly in shrimp in captivity, all marine crustaceans are susceptible. Thus, when the host’s defense mechanisms are suppressed (as is the case when shrimp experience certain environmental triggers), V. harveyi becomes an opportunistic pathogen and releases exotoxins (that destroy the cell or alter the cell’s metabolism). The bacteria has been determined to be ingested with food eaten by the shrimp, but are difficult to eliminate. The bacteria are quite resistant to chlorination and lime treatment to the captivated shrimps’ habitat.[4] The effects of the disease caused by the bioluminescent bacteria are seen rapidly, so finding a method to curtail or eliminate its presence has been difficult given such a short amount of time.
Pathology
Vibrio harveyi has no known pathogenicity to humans, which makes it a good bacteria to study in order to learn more about how bacteria (many of which are pathogenic to humans) function. Since only some strains of V. harveyi are virulent, a large amount of genetic diversity within the bacteria is likely; this characteristic may also increase the difficulty of controlling their virulent effects. V. harveyi, as one of the bacteria responsible for causing vibriosis in shell fish and finfish, may have effects in any of the following manners of the host organism: oral, enteric, appendage, open wounds, systemic, sepsis, and the shell. In shrimp, symptoms may present as: loss of limb function, muscular cloudiness, lesions, gastrointestinal infections, discoloration in the gills, detachment of epithelial cells from the basal ganglia (which is physiologically similar to a disease seen in humans called Bullous pemphigoid). None of these symptoms have been observed in shrimp not infected by V. harveyi.[4]
At least for a certain strain of Vibrio harveyi (VH1114), the bacteria is thought to demonstrate a type of pseudolysogeny. That is, VH1114 proceeds throught the steps of lysogeny, but also undergoes chromosomal alteration from that of the parent chromosome. This part is thought to occur by means of an episome (a type of plasmid that is a component in all V. harveyi genomes, but which is not essential to the replication of the chromosome).[8]
Application to Biotechnology
Understanding the mechanisms by which Vibrio harveyi communicate with their environment and establish bioluminescence(as well as other bacteria of the vibrionaceae family) can help to develop chemicals, compounds, and drugs that that can control (inhibit, in most cases) these pathways and thus control or prevent the virulent effects of V. harveyi. If this can be done, than perhaps some bacterial infections and economically consequential crop failures can be remedied.
Current Research
The Bassler Lab at Princeton University has determined that Vibrio harveyi uses quorum sensing through AI-1(intra-species) and AI-2 (inter-species) circuits, which allow it to function as a multicellular organism. With this knowledge, they are now trying to develop molecules that use AI-2 circuits because these molecules will have the potential to further develop anti-microbial drugs that control virulence. These drugs would hopefully be able to be applied to many bacterial strains, some of which may even be pathogenic to humans.[8]
Recent studies have indicated the use of Vibrio harveyi as an additional indicator of death caused by drowning in humans. Many bioluminescent bacteria were found in blood samples of cadavers that had been immersed in water. Blood from both sides of the heart and the femoral vein contained bioluminescent bacteria—one of which was V. harveyi—when cultured on agar plates. This finding may help to support a conclusion of death by drowning when used in conjunction with police reports and other autopsy findings.[9]
Another current area of research involving V. harveyi is that of vaccine development. The outer membrane proteins of V. harveyi have a role in the interaction between them (the bacterium) and their host. The gene encoding for this outer protein, OmpK, was subcloned and a recombinant protein was found in substantial amounts (24.8%). Fish who were then injected with these recombinant OmpK proteins developed antibodies against V. harveyi. Thus, immunity to V. harveyi by marine organisms may be possible and is further being studied.[10]
References
- ↑ Classification. (2007). Vibrio harveyi. Retrieved April 10, 2009, from Microbe Wiki Web site:http://microbewiki.kenyon.edu/index.php/Vibrio_harveyi
- ↑ 2.0 2.1 Vibrio harveyi genome [Data file]. (n.d.). Retrieved April 18, 2009, from Kegg Web site:http://www.genome.jp/kegg-bin/show_organism?org=vha
- ↑ 3.0 3.1 3.2 3.3 Background. (n.d.). Vibrio harveyi. Retrieved April 10, 2009, from Washington University in St. Louis School of Medicine Genome Sequencing Center Web site: http://genome.wustl.edu/genome.cgl?GENOME=Vibrio%20harveyi
- ↑ 4.0 4.1 4.2 Rao, A. V. (2008, September 12). Vibriosis in shrimp aquaculture. Retrieved April 10, 2009, from http://www.engormix/e_articles_view.asp?art=1179&AREA=ACU-165
- ↑ 5.0 5.1 5.2 Bassler, B. (n.d.). Cell-to-cell communication in bacteria. Retrieved April 10, 2009, from Department of Molecular Biology Princeton University Web site:http://www.molbio.princeton.edu/index.php?option=content&task=view&id=27
- ↑ Pompeanl, A. J., Irgon, J. J., Berger, M. F., Bulyk, M. L., Wingreen, N. S., & Bassler, B. L. (2008, August 15). The vibrio harveyi master quorum-sensing regulator, luxR, a tetR-type protein is both an activator and repressor: DNA recognition and binding specificity at target promoters.Molecular Microbiology, 70(1), 76-88. Retrieved April 10, 2009. doi:10.1111/ j.1365-2958.2008.06389.x
- ↑ Lenz, D. H., Mok, K. C., Lilley, B. N., Kulkarni, R. V., Wingreen, N. S., & Bassler, B. L. (2004,July 9). The small rna chaperone hfq and multiple small rnas control quorum sensing in vibrio harveyi and vibrio cholerae. Cell, 118(1), 69-82. Abstract retrieved April 13, 2009, from Laboratory of Molecule Biology, National Cancer Institute Web site: http://lib.bioinfo.pl/pmld:15242645
- ↑ 8.0 8.1 Khemayan, K., Pasharawipas, T., Puiprom, O., Sriurairatana, S., Suthienkul, O., & Flegel, T. W. (2006, February). Unstable lysogeny and pseudolysogeny in vibrio harveyi siphovirus-like phage 1. Applied Environmental Microbiology, 72(2), 1355-1363. Retrieved April 10, 2009. doi:10.1128/AEM.72.2.1355-1363.2006
- ↑ Kakizaki, E., Kozawa, S., Sakai, M., & Yukawa, N. (2009, January 3). Bioluminescent bacteria have potential as a marker of drowning in seawater: Two immersed cadavers retrieved near estuaries. Legal Medicine (Tokyo), 11(2), 91-96. Abstract retrieved April 29, 2009, from http://www.ncbi.nlm.nih.gov/pubmed/19121971?ordinalpos=16&itool=EntrezSystem2.PEntrez.Pubmed.Pubmed_ResultsPanel.Pubmed_DefaultReportPanel.Pubmed_RVDocSum
- ↑ Ninggiu, L., Junjie, B., Xiaozhe, F., Xing, Y., & Cunbin, S. (2008, September 23). An outer membrane protein, OmpK, is an effective vaccine candidate for Vibrio harveyi in Orange-spotted grouper (Epinephelus coioides). Fish and shellfish immunology, 25(6), 829-833. Abstract retrieved April 29, 2009, from Web site: http://www.ncbi.nlm.nih.gov/pubmed/18854216?ordinalpos=31&itool=EntrezSystem2.PEntrez.Pubmed.Pubmed_ResultsPanel.Pubmed_DefaultReportPanel.Pubmed_RVDocSum