Vibrio fischeri: Difference between revisions
imported>Yeong M Shin |
mNo edit summary |
||
| (81 intermediate revisions by 4 users not shown) | |||
| Line 1: | Line 1: | ||
{{subpages}} | |||
{{Taxobox | |||
| color = pink | |||
| name = ''Vibrio fischeri'' | |||
| image = Vibrio_Fischeri-_PNAS_2005;_102(8)_2673-4,_Figure_2.1_lores.jpg | |||
| regnum = [[Bacteria]] | |||
| phylum = [[Proteobacteria]] | |||
| classis = [[Gammaproteobacteria]] | |||
| ordo = [[Vibrionales]] | |||
| familia = [[Vibrionaceae]] | |||
| genus = ''[[Aliivibrio]]'' | |||
| species = ''[[V. fischeri]]'' | |||
| binomial = ''[[Vibrio fischeri]]'' | |||
| binomial_authority = | |||
}} | |||
==Description and significance== | |||
'''''Vibrio fischeri''''' is a [[Gram-negative]] [[bioluminescent]] marine bacterium that forms mutually [[symbiotic]] relationships with various species of fish and squids.<ref>Ruby, E. G., McFall-Ngai, M. J. Oxygen-utilizing reactions and symbiotic colonization of the squid light organ by Vibrio fischeri. Trends in microbiology. 415 Volume 7, issue 10, October 1999, Pages 414-420. Accessed from ScienceDirect.</ref> ''V. fischeri'' is a member of the Vibrionacea family of [[marine γ-proteobacteria]] which includes many species that have evolved both beneficial and harmful relationships with animals.<ref>Ruby, E. G., M. Urbanowski, J. Campbell, A. Dunn, M. Faini, R. Gunsalus, P. Lostroh, C. Lupp, J. McCann, D. Millikan, A. Schaefer, E. Stabb, A. Stevens, K. Visick, C. Whistler, and E. P. Greenberg. 2005. Complete genome sequence of Vibrio fischeri: a symbiotic bacterium with pathogenic congeners. Proc. Natl. Acad. Sci. USA 102:3004-3009</ref> The motile bacterium can be found living freely in the oceans or inside the microhabitats of its host's light organs. The proteins necessary for the production of bioluminescence are encoded in a set of genes called the ''lux'' [[operon]]. The bioluminescent bacteria produces light in a chemical reaction where [[luciferin]], a substrate molecule, is oxidized by an enzyme, [[luciferase]]. As a result, energy in the form of blue-green light (480-490nm) is emitted.<ref>Herring, P.J. and Widder, E.A.2001. Bioluminescence in Plankton and Nekton. In: Steele, J.H., Thorpe, S.A. and Turekian, K.K. editors, Encyclopedia of Ocean Science, Vol. 1, 308-317. Academic Press, San Diego. Accessed from "International society of bioluminescence and chemiluminescence", <http://www.isbc.unibo.it/Files/BC_PlanktonNekton.htm> (updated 10/2009; accessed 03/20/09). </ref> | |||
Bioluminescence and other metabolic functions in ''V. fischeri'' are regulated by a cell density dependent system called quorum sensing. Quorum sensing occurs through the production and accumulation of a signal molecule, [[N-acyl homoserine lactone]] (AHL); as a consequence individual bacteria do not luminesce. Since its discovery in ''V. fischeri'', quorum sensing and it's respective signal molecules have been shown to regulate a variety of genes whose proteins are required for virulence factors, symbiosis, biofilm formation, plasmid transfer and morphogenesis in many Gram-negative and Gram-positive bacteria.<ref>Willey, J.M., Sherwood, L.M., Woolverton, C. J. Prescott, Harley, and Klein's Microbiology. 7th Ed. 2008. McGraw-Hill. New York. | |||
</ref>. <br/> | |||
The symbiotic relationship between a strain of'' V. fischeri'' and its host, the Hawaiian bobtail squid ''Euprymna scolopes'', has been studied extensively and represents a model system of bacteria/animal symbiosis. Comparative studies are examining this symbiosis model and pathogenic members within the genus vibrios, to detect patterns and similarities in bacteria/host relationships. The information may provide insights into the mechanisms behind pathogenesis and pathogenic/host relationships. <br/> | |||
The isolation and cloning of the lux genes from ''V. fischeri'', and their use as a reporter, have provided scientists with another valuable visual tool to examine living organisms at the cellular level. Similarly, ''V. fischeri'' cells have been made commercially available and are used in the field of ecotoxicology to detect contaminants in the environment more quickly and cheaply than conventional methods. | |||
==Quorum sensing== | |||
Through the use of signal molecules, quorum sensing confers the ability for bacteria to detect one another and to regulate metabolic functions in a cell density dependent manner. Multiple quorum sensing molecules have been discovered in the ''V. fischeri'' isolates of ''E. scolopes'' that function to regulate bioluminescence and other genes during different stages of cell growth that correspond to differences in cell densities. <ref> Lupp,C. and Ruby, E.G. Vibrio fischeri uses two quorum sensing systems for the regulation of early and late colonization factors. Journal of Bacteriology. June 2005, p.3620-3629. Vol.187:11.</ref><br/> | |||
The first AHL signal molecule to be recognized was N-(3-oxohexanoyl) homoserine lactone, produced by an enzyme encoded by the luxI gene. The product of the luxI gene induces bioluminescence genes luxICDABEG during late stage exponential growth, when cell densities are high. The luxI signal molecule is an autoinducer because it forms a positive feedback loop by transcribing its own genes. Bioluminescence occurs when the luxI signal molecule diffuses freely across the plasma membrane into the extracellular environment and accumulates with increasing cell numbers. At a critical population threshold, the concentration of luxI signal molecule is greater outside than inside the cell and the difference in the concentration gradient causes the luxI signal molecule to diffuse back into the cell. Once this happens, the signal molecule binds to a membrane bound transcriptional activator protein encoded by the luxR gene. The luxR and luxI products form a transcriptional complex that binds to the promoter region known as the lux box and transcription of the bioluminescence gene luxICDABEG, occurs. <ref>Willey, J.M., Sherwood, L.M., Woolverton, C. J. Prescott, Harley, and Klein's Microbiology. 7th Ed. 2008. McGraw-Hill. New York.</ref><br/> | |||
A second signal molecule, also a AHL derivative N-(octanoyl) homoserine lactone produced by an enzyme encoded in the ainS gene, regulates many genes including bioluminescence genes during early growth periods consistent with lower cell densities and regulatory genes that promote late stage quorum sensing process. The ainS signal molecule induces late stage quorum sensing by deactivation a negative regulator of the litR gene. LitR is a positive regulator that promotes the transcription of luxR. The ainS signal molecule can also directly bind to the transcriptional activator luxR to produce bioluminescence, although the emission is weaker.<ref> Lupp,C. and Ruby, E.G. Vibrio fischeri uses two quorum sensing systems for the regulation of early and late colonization factors. Journal of Bacteriology. June 2005, p.3620-3629. Vol.187:11.</ref> | |||
Separate mutational analysis of the ainS and luxI genes have shown that these quorum sensing signal molecules are essential for a successful colonization by V. fischeri to its host. Current research has shown that the ainS signal molecule also regulates motility genes and many other genes that have yet to be characterized. | |||
==Bioluminescence== | |||
Bioluminescence is the ability of living organisms to produce light. It has evolved independently multiple times and occurs in a variety of marine species as well as some terrestrial forms. There are various mechanisms leading to light production and there are many different forms of luciferins and luciferases. However, all bioluminescence reactions require the use of molecular oxygen <ref>Widder, E. A. "Marine Bioluminescence." Harbor Branch Oceanographic Institution. <http://gupea.ub.gu.se/dspace/bitstream/2077/19437/5/gupea_2077_19437_5.pdf> (created 2001; accessed 03/28/09)</ref>.<br/> | |||
The bacterial luciferase is a multifunctional enzyme, required for both bioluminescence and aerobic respiration reactions. In the bioluminescence reaction, luciferase catalyses the dual oxidation of luciferin, a reduced riboflavin mononucleotide (FMNH<sub>2</sub>) and an associated molecule, a long chain aldehyde. ATP is required to catalyze this reaction and the resulting products are light, water, oxyluciferin and carboxyl group<ref>Ruby, E. G., McFall-Ngai, M. J. Oxygen-utilizing reactions and symbiotic colonization of the squid light organ by Vibrio fischeri. Trends in microbiology. 415 Volume 7, issue , 10 October 1999, Pages 414-420. Accessed from ScienceDirect.</ref>. | |||
The luciferase enzyme is a heterodimer consisting of alpha and beta subunits, encoded by genes luxA and luxB. LuxCDE form the fatty acid reductase complex, the enzymes required for the production of the long chain fatty aldehyde. The enzyme required to reduce the flavin mononucleotide is encoded by the gene luxG.<ref> Meighen, E.A. Genetics of bacterial bioluminescence. Annual Review of Genetics. Vol.28:117-139. Dec 1994.</ref> | |||
{{Image|juvenile_squid1a.jpg|left|250px|Juvenile ''E. scolopes'', symbiotic host of ''V. fischeri''. Adult bobtail squids grow to a maximum size of 35mm.}} | |||
==Ecology== | ==Ecology== | ||
''V. | |||
Outside from its primary hosts, ''V. fischeri'' can be found living freely as ‘marine snow’, in fecal pellets, as saprohytes <ref>Herring, P.J. and Widder, E.A.2001. Bioluminescence in Plankton and Nekton. In: Steele, J.H., Thorpe, S.A. and Turekian, K.K. editors, Encyclopedia of Ocean Science, Vol. 1, 308-317. Academic Press, San Diego. Accessed from "International society of bioluminescence and chemiluminescence", <http://www.isbc.unibo.it/Files/BC_PlanktonNekton.htm> (updated 10/2009; accessed 03/20/09</ref>, and amongst the microbial flora in the guts of marine animals. The heterotrophic bacteria are distributed in the pelagic zone of temperate and subtropical waters <ref>Ruby, E. G., McFall-Ngai, M. J. Oxygen-utilizing reactions and symbiotic colonization of the squid light organ by Vibrio fischeri. Trends in microbiology. 415 Volume 7, issue , 10 October 1999, Pages 414-420. Accessed from ScienceDirect</ref>. | |||
===Symbiosis=== | |||
''E. scolopes'' and ''V. fischeri'' strain ES114 have adaptively coevolved to maintain an exclusive relationship. <ref>Thompson, F.L., Austin, B., Swings, J. The biology of Vibrios. 2006. ASM press. Virginia.</ref> In this symbiotic relationship, ''E.scolopes'' benefits by using the bioluminescent bacteria in a camouflage strategy called ‘counter-illumination’. As the nocturnal squid swims on the surface of the ocean, it projects the light produced by the bacteria ventrally. This effectively mimics moonlight when viewed from below and the squid projects no shadow that can be detected by a predator. ''E. scolopes'' regulates the amount of bacterial illumination with the light organ and its surrounding tissue. In order for the squid's light organ to develop, the organ must be inoculated with the bacteria; otherwise, the viability of ''E. scolopes'' in the wild is greatly diminished. In exchange, ''V. fischeri'' is provided housing and nutrients within the crypts of the light organ. Once colonized within the light organ, the bacterial population can increase to 10<sup>9</sup>-10<sup>11</sup> cells, of which 90-95% are expelled every morning by the squid. Th remaining bacteria repopulate the light organ, and the squid becomes a vehicle for bacterial proliferation. <ref>Ruby, E. G., McFall-Ngai, M. J. Oxygen-utilizing reactions and symbiotic colonization of the squid light organ by Vibrio fischeri. Trends in microbiology. 415 Volume 7, issue , 10 October 1999, Pages 414-420. Accessed from ScienceDirect</ref><br/> | |||
Newly hatched squids are born without the bacterium and must acquire them from the surrounding seawaters. ''V. fischeri'' may constitute only 0.1% of total microbes per mL of seawater, yet the squid becomes inoculated with ''V. fischeri'' within a few minutes after its birth. Furthermore, results from an experiment using different strains of ''V. fischeri'' to inoculate ''E. scolopes'' have shown that non-native strains of ''V. fischeri'' cannot colonize the host as effectively as the native strain. <ref>McFall-Ngai, M. J. 2000. Negotiations between animals and bacteria: the 'diplomacy' of the squid-vibrio symbiosis. Comp. Biochem. Physiol. A. Mol. Integr. Physiol., 126(4), 471-480.</ref><br/> | |||
There are two important aspects in the selective colonization of the squid by ''V. fischeri''. The squid inhibits unwanted bacteria by producing reactive oxygen species (ROS). Also, the interaction between ''V. fischeri'' and host cells induces differential gene expression in each other which are required for a successful colonization. <ref>Ruby, E. G., McFall-Ngai, M. J. Oxygen-utilizing reactions and symbiotic colonization of the squid light organ by Vibrio fischeri. Trends in microbiology. 415 Volume 7, issue , 10 October 1999, Pages 414-420. Accessed from ScienceDirect</ref> <br/> | |||
In the infection stage of the colonization process, free floating bacteria in the ocean induces the newly hatched squid to secrete mucus from an appendage of the light organ known as the ciliated fields. It is thought that chemotaxis may be involved in the movement of ''V. fischeri'' towards the squid mucus. ''V. fischeri'' has been shown to be highly attracted to nucleosides and N-acetylneuraminic, which are components of the squid mucus. ''V. fischeri'' and other bacteria adhere to the mucus, however, nitric oxide (a ROS) contained in the mucus limits unwanted bacteria while promoting the symbionts <ref>Thompson, F.L., Austin, B., Swings, J. The biology of Vibrios. 2006. ASM press. Virginia.</ref><br/> | |||
In the next stage, ''V. fischeri'' must use its flagella to migrate from the mucus aggregation to pores leading into the crypts of the light organ. Narrow ciliated ducts connect the pores to the light organ. Motility mutants with overexpressed or non-functional flagella genes were unable to colonize ''E. scolopes'' in the initial stages. <ref>Visick, K., Ruby, E. G. Vibrio fischeri and its host: it takes two to tango. Current Opinion in Microbiology. Volume 9, Issue 6, December 2006, Pages 632-638. Accessed from ScienceDirect.</ref> | |||
<br/> | |||
The interior of the light organ has been shown to be a dynamic and complex environment where ''V. fischeri'' and host cells interact. <ref>Thompson, F.L., Austin, B., Swings, J. The biology of Vibrios. 2006. ASM press. Virginia.</ref> The crypt of the light organ contains halide peroxidase (HPO), another ROS. However, colonization by ''V. fischeri'' has been shown to lower the mRNA of HPO, resulting in the decreased production of HPO. <ref>Ruby, E. G., McFall-Ngai, M. J. Oxygen-utilizing reactions and symbiotic colonization of the squid light organ by Vibrio fischeri. Trends in microbiology. 415 Volume 7, issue , 10 October 1999, Pages 414-420. Accessed from ScienceDirect</ref>Colonization of the crypts by the bacteria also induces morphogenesis for both the bacterium and its host. ''V. fischeri'' secretes a compound identical to a tracial cytotoxin (TCT) produced by the pathogen ''Bordetella pertussis'', peptidoglycan tetrapeptide monomer, that induces normal apoptosis of the ciliated fields. Concurrently, the light organ swells with increased microvilli production. ''V. fischer''i, on the other hand, lose their flagella and cell size decreases.<ref>Visick, K., Ruby, E. G. Vibrio fischeri and its host: it takes two to tango. Current Opinion in Microbiology. Volume 9, Issue 6, December 2006, Pages 632-638. Accessed from ScienceDirect.</ref> | |||
{{Image|JuvScolopesDiagram.jpg|center|500px|Inoculation pathway of ''V. fischeri'' into light organ of juvenile ''E. scolopes''. Figure a- the blue lines and arrows depict the movement of "V. fischeri" cells from surrounding seawater into the mantle cavity of newly hatched "E. Scolopes" (white outline). As the water travels into the cavity's funnel, it interacts with the developing light organ, depicted by the green-hued laser-scanning confocal micrograph (LSM) in the illustration. The bright green areas of the organ are complex ciliated fields. Figure b- This LSM shows pores at the base of the ciliated fields of the light organ.}}<ref> Nyholm, S., Stabb, E., Ruby, E. and McFall-Ngai, M. Establishment of an animal-bacterial association: Recruiting symbiotic vibrios from the environment. PNAS. August 29, 2000. Vol.97. no 18, 10231-10235.</ref> | |||
==Genome== | |||
To date, the complete genomes of two strains of ''V. fischeri'', ES114 and MJII have been sequenced. MJ11 is isolated from the japanese pinecone fish, ''Monocentris japonica''. Strain ES114 isolated from ''E.scolopes'' contains two circular chromosomes and a circular plasmid pES100. The genetic material is composed of DNA and the total genome is 4.284 Mbp in length. Chromosome 1, the larger of the two chromosomes contains 2586 genes, while Chromosome 2 and the plasmid contains 1175 and 57 genes respectively. | |||
The lux operon system, located on chromosome 2, contains luxI, luxR and luxCDABEG. | |||
<ref>NCBI. Entrez genome project. <http://www.ncbi.nlm.nih.gov/sites/entrez?Db=genomeprj&Cmd=Retrieve&list_uids=12986>(updated 2/09;accessed 03/29/09)</ref>. | |||
==Pathology== | |||
''V. fischeri'' is non-pathogenic to humans but is pathogenic to some marine invertebrates.<ref>Ruby, E. G., McFall-Ngai, M. J. Oxygen-utilizing reactions and symbiotic colonization of the squid light organ by Vibrio fischeri. Trends in microbiology. 415 Volume 7, issue , 10 October 1999, Pages 414-420. Accessed from ScienceDirect</ref> Within the genus Vibrios, there are three human pathogens ''V. cholerae'', ''V.parahaemolytus'', ''V. vulnificus''. | |||
==Application to Biotechnology== | |||
The lux genes isolated from various bioluminescent organisms, in combination with the use of recombinant DNA technology, have had wide applications in the field of science. One of the primary uses of bioluminescent genes is as a reporter gene. A reporter gene allows scientists to visually track proteins and molecular processes occurring inside of living organisms. Scientists use reporter genes to uncover various characteristics of proteins, determine the functions of genes, and to describe regulatory regions of the genome.<br/> | |||
The ''V. fischeri'' bacteria and its isolated lux operon are being used to detect pollution in the environment. In using the whole-cell approach, non-specific contamination is detected by the decrease in cell luminescence brought upon by cell death or metabolic failure. Methods to detect for specific contaminants utilize the lux operon and a regulatory region that is specific to the target compound.<ref> Tamminen, M.V. and Virta, M., Quantification of ecotoxicological tests based on bioluminescence using polaroid film. Chemosphere 66 (2007) 1329-1335. Accessed from Science Direct.</ref> | |||
==Current Research== | |||
In recent line of research, Anderson and colleagues engineered bacteria that are able to target and invade mammalian cancer cells in part using ''V. fischeri'' quorum sensing genes. The luxI-luxR quorum sensing genes were used as a regulatory sensor to target cancer cells because bacteria have been shown to localize on tumor afflicted regions in numbers greater than 10<sup>9</sup> when injected intravenously. Bacteria are attracted to cancer cells due to lack of immune surveillance, as well as the hypoxic and nutrient-rich environment. The density dependent quorum sensing system triggers the invasin gene only with high cell numbers. The invasin gene, isolated from ''Yersinia pseudotuberculosis'', allows bacteria to adhere to and invade mammalian cells by binding to β1-integrin surface proteins. With this bacteria construction, the scientist are in the process of developing ways to deliver proteins and therapies to the cancer cells. <ref>Anderson,C., Clarke, E..J., Arkin, A.P., Voigt, C.A. Environmentally controlled invasion of cancer cells by engineered bacteria. Journal of Molecular Biology. 2006. 355;619-627. Accessed from Science Direct 4/23/09 </ref><br/> | |||
Mandel and colleagues found that a single regulatory gene, rscS in ''V. fischeri'', was necessary for establishment for symbiosis by strain ES114 to its host ''E.scolopes''. Comparative genomic studies of the rscS gene in different strains of ''V. fischeri'' isolated from squid and fish hosts revealed that the squid isolates all had a conserved rscS gene. From the fish isolates, five out of the ten fish also had a conserved rscS gene, but highly divergent from the squid isolates. In the experiment, the rscs gene was inserted into the MJ11 strains isolated from fish that cannot effectively colonize squid. With the transformation, the MJ11 strains were able to colonize the light organ as effectively as the natural symbionts. <ref>Mandel, M.J., Wollenberg, M.S., Stabb, E.V.,Visick, K.L.,Ruby, E.G. A single regulatory gene is sufficient to alter bacterial host range. Nature. 2009 Mar 12;458(7235):215-8.</ref> | |||
==References== | |||
{{reflist|2}} | |||
*Anderson,C., Clarke, E..J., Arkin, A.P., Voigt, C.A. Environmentally controlled invasion of cancer cells by engineered bacteria. Journal of Molecular Biology. 2006. 355;619-627. Accessed from Science Direct 4/23/09 | |||
*Fuqua,C., Winans,S., Greenberg, P. Census and consensus in bacterial ecosystems: the luxR-luxI family of quorum-sensing transciptional regulators. Annual Review of Microbiology. Vol. 50: 727-751 (Volume publication date October 1996) | |||
*Herring, P.J. and Widder, E.A.2001. Bioluminescence in Plankton and Nekton. In: Steele, J.H., Thorpe, S.A. and Turekian, K.K. editors, Encyclopedia of Ocean Science, Vol. 1, 308-317. Academic Press, San Diego. Accessed from "International society of bioluminescence and chemiluminescence", <http://www.isbc.unibo.it/Files/BC_PlanktonNekton.htm> (updated 10/2009; accessed 03/20/09) | |||
*Mandel, M.J., Wollenberg, M.S., Stabb, E.V.,Visick, K.L.,Ruby, E.G. A single regulatory gene is sufficient to alter bacterial host range. Nature. (In press) | |||
*McFall-Ngai, M. J. 2000. Negotiations between animals and bacteria: the 'diplomacy' of the squid-vibrio symbiosis. Comp. Biochem. Physiol. A. Mol. Integr. Physiol., 126(4), 471-480. | |||
*Meighen, E.A. Genetics of bacterial bioluminescence. Annual Review of Genetics. Vol.28:117-139. Dec 1994. | |||
*NCBI. Entrez genome project. <http://www.ncbi.nlm.nih.gov/sites/entrez?Db=genomeprj&Cmd=Retrieve&list_uids=12986>(updated 2/09;accessed 03/29/09) | |||
*Nyholm, S., Stabb, E., Ruby, E. and McFall-Ngai, M. Establishment of an animal-bacterial association: Recruiting symbiotic vibrios from the environment. PNAS. August 29, 2000. Vol.97. no 18, 10231-10235 | |||
*Ruby, E. G., M. Urbanowski, J. Campbell, A. Dunn, M. Faini, R. Gunsalus, P. Lostroh, C. Lupp, J. McCann, D. Millikan, A. Schaefer, E. Stabb, A. Stevens, K. Visick, C. Whistler, and E. P. Greenberg. 2005. Complete genome sequence of Vibrio fischeri: a symbiotic bacterium with pathogenic congeners. Proc. Natl. Acad. Sci. USA 102:3004-3009 | |||
*Ruby, E. G., McFall-Ngai, M. J. Oxygen-utilizing reactions and symbiotic colonization of the squid light organ by Vibrio fischeri. Trends in microbiology. 415 Volume 7, issue , 10October 1999, Pages 414-420. Accessed from ScienceDirect | |||
*Tamminen, M.V. and Virta, M., Quantification of ecotoxicological tests based on bioluminescence using polaroid film. Chemosphere 66 (2007) 1329-1335. Accessed from Science Direct | |||
*Thompson, F.L., Austin, B., Swings, J. The biology of Vibrios. 2006. ASM press. Virginia. | |||
*Visick, K., Ruby, E. G. Vibrio fischeri and its host: it takes two to tango. Current Opinion in Microbiology. Volume 9, Issue 6, December 2006, Pages 632-638. Accessed from ScienceDirect. | |||
*Willey, J.M., Sherwood, L.M., Woolverton, C. J. Prescott, Harley, and Klein's Microbiology. 7th Ed. 2008. McGraw-Hill. New York. | |||
[[Category:Suggestion Bot Tag]] | |||
Latest revision as of 17:01, 4 November 2024
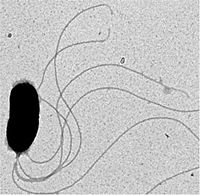
| Vibrio fischeri | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
 | ||||||||||||||
| Scientific classification | ||||||||||||||
| ||||||||||||||
| Binomial name | ||||||||||||||
| Vibrio fischeri |
Description and significance
Vibrio fischeri is a Gram-negative bioluminescent marine bacterium that forms mutually symbiotic relationships with various species of fish and squids.[1] V. fischeri is a member of the Vibrionacea family of marine γ-proteobacteria which includes many species that have evolved both beneficial and harmful relationships with animals.[2] The motile bacterium can be found living freely in the oceans or inside the microhabitats of its host's light organs. The proteins necessary for the production of bioluminescence are encoded in a set of genes called the lux operon. The bioluminescent bacteria produces light in a chemical reaction where luciferin, a substrate molecule, is oxidized by an enzyme, luciferase. As a result, energy in the form of blue-green light (480-490nm) is emitted.[3]
Bioluminescence and other metabolic functions in V. fischeri are regulated by a cell density dependent system called quorum sensing. Quorum sensing occurs through the production and accumulation of a signal molecule, N-acyl homoserine lactone (AHL); as a consequence individual bacteria do not luminesce. Since its discovery in V. fischeri, quorum sensing and it's respective signal molecules have been shown to regulate a variety of genes whose proteins are required for virulence factors, symbiosis, biofilm formation, plasmid transfer and morphogenesis in many Gram-negative and Gram-positive bacteria.[4].
The symbiotic relationship between a strain of V. fischeri and its host, the Hawaiian bobtail squid Euprymna scolopes, has been studied extensively and represents a model system of bacteria/animal symbiosis. Comparative studies are examining this symbiosis model and pathogenic members within the genus vibrios, to detect patterns and similarities in bacteria/host relationships. The information may provide insights into the mechanisms behind pathogenesis and pathogenic/host relationships.
The isolation and cloning of the lux genes from V. fischeri, and their use as a reporter, have provided scientists with another valuable visual tool to examine living organisms at the cellular level. Similarly, V. fischeri cells have been made commercially available and are used in the field of ecotoxicology to detect contaminants in the environment more quickly and cheaply than conventional methods.
Quorum sensing
Through the use of signal molecules, quorum sensing confers the ability for bacteria to detect one another and to regulate metabolic functions in a cell density dependent manner. Multiple quorum sensing molecules have been discovered in the V. fischeri isolates of E. scolopes that function to regulate bioluminescence and other genes during different stages of cell growth that correspond to differences in cell densities. [5]
The first AHL signal molecule to be recognized was N-(3-oxohexanoyl) homoserine lactone, produced by an enzyme encoded by the luxI gene. The product of the luxI gene induces bioluminescence genes luxICDABEG during late stage exponential growth, when cell densities are high. The luxI signal molecule is an autoinducer because it forms a positive feedback loop by transcribing its own genes. Bioluminescence occurs when the luxI signal molecule diffuses freely across the plasma membrane into the extracellular environment and accumulates with increasing cell numbers. At a critical population threshold, the concentration of luxI signal molecule is greater outside than inside the cell and the difference in the concentration gradient causes the luxI signal molecule to diffuse back into the cell. Once this happens, the signal molecule binds to a membrane bound transcriptional activator protein encoded by the luxR gene. The luxR and luxI products form a transcriptional complex that binds to the promoter region known as the lux box and transcription of the bioluminescence gene luxICDABEG, occurs. [6]
A second signal molecule, also a AHL derivative N-(octanoyl) homoserine lactone produced by an enzyme encoded in the ainS gene, regulates many genes including bioluminescence genes during early growth periods consistent with lower cell densities and regulatory genes that promote late stage quorum sensing process. The ainS signal molecule induces late stage quorum sensing by deactivation a negative regulator of the litR gene. LitR is a positive regulator that promotes the transcription of luxR. The ainS signal molecule can also directly bind to the transcriptional activator luxR to produce bioluminescence, although the emission is weaker.[7]
Separate mutational analysis of the ainS and luxI genes have shown that these quorum sensing signal molecules are essential for a successful colonization by V. fischeri to its host. Current research has shown that the ainS signal molecule also regulates motility genes and many other genes that have yet to be characterized.
Bioluminescence
Bioluminescence is the ability of living organisms to produce light. It has evolved independently multiple times and occurs in a variety of marine species as well as some terrestrial forms. There are various mechanisms leading to light production and there are many different forms of luciferins and luciferases. However, all bioluminescence reactions require the use of molecular oxygen [8].
The bacterial luciferase is a multifunctional enzyme, required for both bioluminescence and aerobic respiration reactions. In the bioluminescence reaction, luciferase catalyses the dual oxidation of luciferin, a reduced riboflavin mononucleotide (FMNH2) and an associated molecule, a long chain aldehyde. ATP is required to catalyze this reaction and the resulting products are light, water, oxyluciferin and carboxyl group[9].
The luciferase enzyme is a heterodimer consisting of alpha and beta subunits, encoded by genes luxA and luxB. LuxCDE form the fatty acid reductase complex, the enzymes required for the production of the long chain fatty aldehyde. The enzyme required to reduce the flavin mononucleotide is encoded by the gene luxG.[10]
Ecology
Outside from its primary hosts, V. fischeri can be found living freely as ‘marine snow’, in fecal pellets, as saprohytes [11], and amongst the microbial flora in the guts of marine animals. The heterotrophic bacteria are distributed in the pelagic zone of temperate and subtropical waters [12].
Symbiosis
E. scolopes and V. fischeri strain ES114 have adaptively coevolved to maintain an exclusive relationship. [13] In this symbiotic relationship, E.scolopes benefits by using the bioluminescent bacteria in a camouflage strategy called ‘counter-illumination’. As the nocturnal squid swims on the surface of the ocean, it projects the light produced by the bacteria ventrally. This effectively mimics moonlight when viewed from below and the squid projects no shadow that can be detected by a predator. E. scolopes regulates the amount of bacterial illumination with the light organ and its surrounding tissue. In order for the squid's light organ to develop, the organ must be inoculated with the bacteria; otherwise, the viability of E. scolopes in the wild is greatly diminished. In exchange, V. fischeri is provided housing and nutrients within the crypts of the light organ. Once colonized within the light organ, the bacterial population can increase to 109-1011 cells, of which 90-95% are expelled every morning by the squid. Th remaining bacteria repopulate the light organ, and the squid becomes a vehicle for bacterial proliferation. [14]
Newly hatched squids are born without the bacterium and must acquire them from the surrounding seawaters. V. fischeri may constitute only 0.1% of total microbes per mL of seawater, yet the squid becomes inoculated with V. fischeri within a few minutes after its birth. Furthermore, results from an experiment using different strains of V. fischeri to inoculate E. scolopes have shown that non-native strains of V. fischeri cannot colonize the host as effectively as the native strain. [15]
There are two important aspects in the selective colonization of the squid by V. fischeri. The squid inhibits unwanted bacteria by producing reactive oxygen species (ROS). Also, the interaction between V. fischeri and host cells induces differential gene expression in each other which are required for a successful colonization. [16]
In the infection stage of the colonization process, free floating bacteria in the ocean induces the newly hatched squid to secrete mucus from an appendage of the light organ known as the ciliated fields. It is thought that chemotaxis may be involved in the movement of V. fischeri towards the squid mucus. V. fischeri has been shown to be highly attracted to nucleosides and N-acetylneuraminic, which are components of the squid mucus. V. fischeri and other bacteria adhere to the mucus, however, nitric oxide (a ROS) contained in the mucus limits unwanted bacteria while promoting the symbionts [17]
In the next stage, V. fischeri must use its flagella to migrate from the mucus aggregation to pores leading into the crypts of the light organ. Narrow ciliated ducts connect the pores to the light organ. Motility mutants with overexpressed or non-functional flagella genes were unable to colonize E. scolopes in the initial stages. [18]
The interior of the light organ has been shown to be a dynamic and complex environment where V. fischeri and host cells interact. [19] The crypt of the light organ contains halide peroxidase (HPO), another ROS. However, colonization by V. fischeri has been shown to lower the mRNA of HPO, resulting in the decreased production of HPO. [20]Colonization of the crypts by the bacteria also induces morphogenesis for both the bacterium and its host. V. fischeri secretes a compound identical to a tracial cytotoxin (TCT) produced by the pathogen Bordetella pertussis, peptidoglycan tetrapeptide monomer, that induces normal apoptosis of the ciliated fields. Concurrently, the light organ swells with increased microvilli production. V. fischeri, on the other hand, lose their flagella and cell size decreases.[21]
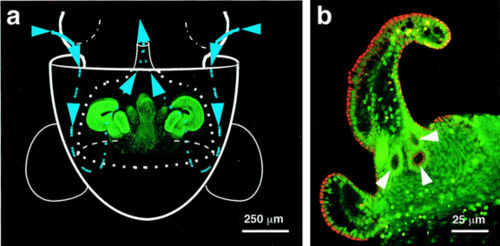
Genome
To date, the complete genomes of two strains of V. fischeri, ES114 and MJII have been sequenced. MJ11 is isolated from the japanese pinecone fish, Monocentris japonica. Strain ES114 isolated from E.scolopes contains two circular chromosomes and a circular plasmid pES100. The genetic material is composed of DNA and the total genome is 4.284 Mbp in length. Chromosome 1, the larger of the two chromosomes contains 2586 genes, while Chromosome 2 and the plasmid contains 1175 and 57 genes respectively. The lux operon system, located on chromosome 2, contains luxI, luxR and luxCDABEG. [23].
Pathology
V. fischeri is non-pathogenic to humans but is pathogenic to some marine invertebrates.[24] Within the genus Vibrios, there are three human pathogens V. cholerae, V.parahaemolytus, V. vulnificus.
Application to Biotechnology
The lux genes isolated from various bioluminescent organisms, in combination with the use of recombinant DNA technology, have had wide applications in the field of science. One of the primary uses of bioluminescent genes is as a reporter gene. A reporter gene allows scientists to visually track proteins and molecular processes occurring inside of living organisms. Scientists use reporter genes to uncover various characteristics of proteins, determine the functions of genes, and to describe regulatory regions of the genome.
The V. fischeri bacteria and its isolated lux operon are being used to detect pollution in the environment. In using the whole-cell approach, non-specific contamination is detected by the decrease in cell luminescence brought upon by cell death or metabolic failure. Methods to detect for specific contaminants utilize the lux operon and a regulatory region that is specific to the target compound.[25]
Current Research
In recent line of research, Anderson and colleagues engineered bacteria that are able to target and invade mammalian cancer cells in part using V. fischeri quorum sensing genes. The luxI-luxR quorum sensing genes were used as a regulatory sensor to target cancer cells because bacteria have been shown to localize on tumor afflicted regions in numbers greater than 109 when injected intravenously. Bacteria are attracted to cancer cells due to lack of immune surveillance, as well as the hypoxic and nutrient-rich environment. The density dependent quorum sensing system triggers the invasin gene only with high cell numbers. The invasin gene, isolated from Yersinia pseudotuberculosis, allows bacteria to adhere to and invade mammalian cells by binding to β1-integrin surface proteins. With this bacteria construction, the scientist are in the process of developing ways to deliver proteins and therapies to the cancer cells. [26]
Mandel and colleagues found that a single regulatory gene, rscS in V. fischeri, was necessary for establishment for symbiosis by strain ES114 to its host E.scolopes. Comparative genomic studies of the rscS gene in different strains of V. fischeri isolated from squid and fish hosts revealed that the squid isolates all had a conserved rscS gene. From the fish isolates, five out of the ten fish also had a conserved rscS gene, but highly divergent from the squid isolates. In the experiment, the rscs gene was inserted into the MJ11 strains isolated from fish that cannot effectively colonize squid. With the transformation, the MJ11 strains were able to colonize the light organ as effectively as the natural symbionts. [27]
References
- ↑ Ruby, E. G., McFall-Ngai, M. J. Oxygen-utilizing reactions and symbiotic colonization of the squid light organ by Vibrio fischeri. Trends in microbiology. 415 Volume 7, issue 10, October 1999, Pages 414-420. Accessed from ScienceDirect.
- ↑ Ruby, E. G., M. Urbanowski, J. Campbell, A. Dunn, M. Faini, R. Gunsalus, P. Lostroh, C. Lupp, J. McCann, D. Millikan, A. Schaefer, E. Stabb, A. Stevens, K. Visick, C. Whistler, and E. P. Greenberg. 2005. Complete genome sequence of Vibrio fischeri: a symbiotic bacterium with pathogenic congeners. Proc. Natl. Acad. Sci. USA 102:3004-3009
- ↑ Herring, P.J. and Widder, E.A.2001. Bioluminescence in Plankton and Nekton. In: Steele, J.H., Thorpe, S.A. and Turekian, K.K. editors, Encyclopedia of Ocean Science, Vol. 1, 308-317. Academic Press, San Diego. Accessed from "International society of bioluminescence and chemiluminescence", <http://www.isbc.unibo.it/Files/BC_PlanktonNekton.htm> (updated 10/2009; accessed 03/20/09).
- ↑ Willey, J.M., Sherwood, L.M., Woolverton, C. J. Prescott, Harley, and Klein's Microbiology. 7th Ed. 2008. McGraw-Hill. New York.
- ↑ Lupp,C. and Ruby, E.G. Vibrio fischeri uses two quorum sensing systems for the regulation of early and late colonization factors. Journal of Bacteriology. June 2005, p.3620-3629. Vol.187:11.
- ↑ Willey, J.M., Sherwood, L.M., Woolverton, C. J. Prescott, Harley, and Klein's Microbiology. 7th Ed. 2008. McGraw-Hill. New York.
- ↑ Lupp,C. and Ruby, E.G. Vibrio fischeri uses two quorum sensing systems for the regulation of early and late colonization factors. Journal of Bacteriology. June 2005, p.3620-3629. Vol.187:11.
- ↑ Widder, E. A. "Marine Bioluminescence." Harbor Branch Oceanographic Institution. <http://gupea.ub.gu.se/dspace/bitstream/2077/19437/5/gupea_2077_19437_5.pdf> (created 2001; accessed 03/28/09)
- ↑ Ruby, E. G., McFall-Ngai, M. J. Oxygen-utilizing reactions and symbiotic colonization of the squid light organ by Vibrio fischeri. Trends in microbiology. 415 Volume 7, issue , 10 October 1999, Pages 414-420. Accessed from ScienceDirect.
- ↑ Meighen, E.A. Genetics of bacterial bioluminescence. Annual Review of Genetics. Vol.28:117-139. Dec 1994.
- ↑ Herring, P.J. and Widder, E.A.2001. Bioluminescence in Plankton and Nekton. In: Steele, J.H., Thorpe, S.A. and Turekian, K.K. editors, Encyclopedia of Ocean Science, Vol. 1, 308-317. Academic Press, San Diego. Accessed from "International society of bioluminescence and chemiluminescence", <http://www.isbc.unibo.it/Files/BC_PlanktonNekton.htm> (updated 10/2009; accessed 03/20/09
- ↑ Ruby, E. G., McFall-Ngai, M. J. Oxygen-utilizing reactions and symbiotic colonization of the squid light organ by Vibrio fischeri. Trends in microbiology. 415 Volume 7, issue , 10 October 1999, Pages 414-420. Accessed from ScienceDirect
- ↑ Thompson, F.L., Austin, B., Swings, J. The biology of Vibrios. 2006. ASM press. Virginia.
- ↑ Ruby, E. G., McFall-Ngai, M. J. Oxygen-utilizing reactions and symbiotic colonization of the squid light organ by Vibrio fischeri. Trends in microbiology. 415 Volume 7, issue , 10 October 1999, Pages 414-420. Accessed from ScienceDirect
- ↑ McFall-Ngai, M. J. 2000. Negotiations between animals and bacteria: the 'diplomacy' of the squid-vibrio symbiosis. Comp. Biochem. Physiol. A. Mol. Integr. Physiol., 126(4), 471-480.
- ↑ Ruby, E. G., McFall-Ngai, M. J. Oxygen-utilizing reactions and symbiotic colonization of the squid light organ by Vibrio fischeri. Trends in microbiology. 415 Volume 7, issue , 10 October 1999, Pages 414-420. Accessed from ScienceDirect
- ↑ Thompson, F.L., Austin, B., Swings, J. The biology of Vibrios. 2006. ASM press. Virginia.
- ↑ Visick, K., Ruby, E. G. Vibrio fischeri and its host: it takes two to tango. Current Opinion in Microbiology. Volume 9, Issue 6, December 2006, Pages 632-638. Accessed from ScienceDirect.
- ↑ Thompson, F.L., Austin, B., Swings, J. The biology of Vibrios. 2006. ASM press. Virginia.
- ↑ Ruby, E. G., McFall-Ngai, M. J. Oxygen-utilizing reactions and symbiotic colonization of the squid light organ by Vibrio fischeri. Trends in microbiology. 415 Volume 7, issue , 10 October 1999, Pages 414-420. Accessed from ScienceDirect
- ↑ Visick, K., Ruby, E. G. Vibrio fischeri and its host: it takes two to tango. Current Opinion in Microbiology. Volume 9, Issue 6, December 2006, Pages 632-638. Accessed from ScienceDirect.
- ↑ Nyholm, S., Stabb, E., Ruby, E. and McFall-Ngai, M. Establishment of an animal-bacterial association: Recruiting symbiotic vibrios from the environment. PNAS. August 29, 2000. Vol.97. no 18, 10231-10235.
- ↑ NCBI. Entrez genome project. <http://www.ncbi.nlm.nih.gov/sites/entrez?Db=genomeprj&Cmd=Retrieve&list_uids=12986>(updated 2/09;accessed 03/29/09)
- ↑ Ruby, E. G., McFall-Ngai, M. J. Oxygen-utilizing reactions and symbiotic colonization of the squid light organ by Vibrio fischeri. Trends in microbiology. 415 Volume 7, issue , 10 October 1999, Pages 414-420. Accessed from ScienceDirect
- ↑ Tamminen, M.V. and Virta, M., Quantification of ecotoxicological tests based on bioluminescence using polaroid film. Chemosphere 66 (2007) 1329-1335. Accessed from Science Direct.
- ↑ Anderson,C., Clarke, E..J., Arkin, A.P., Voigt, C.A. Environmentally controlled invasion of cancer cells by engineered bacteria. Journal of Molecular Biology. 2006. 355;619-627. Accessed from Science Direct 4/23/09
- ↑ Mandel, M.J., Wollenberg, M.S., Stabb, E.V.,Visick, K.L.,Ruby, E.G. A single regulatory gene is sufficient to alter bacterial host range. Nature. 2009 Mar 12;458(7235):215-8.
- Anderson,C., Clarke, E..J., Arkin, A.P., Voigt, C.A. Environmentally controlled invasion of cancer cells by engineered bacteria. Journal of Molecular Biology. 2006. 355;619-627. Accessed from Science Direct 4/23/09
- Fuqua,C., Winans,S., Greenberg, P. Census and consensus in bacterial ecosystems: the luxR-luxI family of quorum-sensing transciptional regulators. Annual Review of Microbiology. Vol. 50: 727-751 (Volume publication date October 1996)
- Herring, P.J. and Widder, E.A.2001. Bioluminescence in Plankton and Nekton. In: Steele, J.H., Thorpe, S.A. and Turekian, K.K. editors, Encyclopedia of Ocean Science, Vol. 1, 308-317. Academic Press, San Diego. Accessed from "International society of bioluminescence and chemiluminescence", <http://www.isbc.unibo.it/Files/BC_PlanktonNekton.htm> (updated 10/2009; accessed 03/20/09)
- Mandel, M.J., Wollenberg, M.S., Stabb, E.V.,Visick, K.L.,Ruby, E.G. A single regulatory gene is sufficient to alter bacterial host range. Nature. (In press)
- McFall-Ngai, M. J. 2000. Negotiations between animals and bacteria: the 'diplomacy' of the squid-vibrio symbiosis. Comp. Biochem. Physiol. A. Mol. Integr. Physiol., 126(4), 471-480.
- Meighen, E.A. Genetics of bacterial bioluminescence. Annual Review of Genetics. Vol.28:117-139. Dec 1994.
- NCBI. Entrez genome project. <http://www.ncbi.nlm.nih.gov/sites/entrez?Db=genomeprj&Cmd=Retrieve&list_uids=12986>(updated 2/09;accessed 03/29/09)
- Nyholm, S., Stabb, E., Ruby, E. and McFall-Ngai, M. Establishment of an animal-bacterial association: Recruiting symbiotic vibrios from the environment. PNAS. August 29, 2000. Vol.97. no 18, 10231-10235
- Ruby, E. G., M. Urbanowski, J. Campbell, A. Dunn, M. Faini, R. Gunsalus, P. Lostroh, C. Lupp, J. McCann, D. Millikan, A. Schaefer, E. Stabb, A. Stevens, K. Visick, C. Whistler, and E. P. Greenberg. 2005. Complete genome sequence of Vibrio fischeri: a symbiotic bacterium with pathogenic congeners. Proc. Natl. Acad. Sci. USA 102:3004-3009
- Ruby, E. G., McFall-Ngai, M. J. Oxygen-utilizing reactions and symbiotic colonization of the squid light organ by Vibrio fischeri. Trends in microbiology. 415 Volume 7, issue , 10October 1999, Pages 414-420. Accessed from ScienceDirect
- Tamminen, M.V. and Virta, M., Quantification of ecotoxicological tests based on bioluminescence using polaroid film. Chemosphere 66 (2007) 1329-1335. Accessed from Science Direct
- Thompson, F.L., Austin, B., Swings, J. The biology of Vibrios. 2006. ASM press. Virginia.
- Visick, K., Ruby, E. G. Vibrio fischeri and its host: it takes two to tango. Current Opinion in Microbiology. Volume 9, Issue 6, December 2006, Pages 632-638. Accessed from ScienceDirect.
- Willey, J.M., Sherwood, L.M., Woolverton, C. J. Prescott, Harley, and Klein's Microbiology. 7th Ed. 2008. McGraw-Hill. New York.
