Horizontal gene transfer/Citable Version: Difference between revisions
imported>David Tribe |
imported>David Tribe |
||
| Line 93: | Line 93: | ||
*{{cite journal | author = Woese C, Fox G | title = Phylogenetic structure of the prokaryotic domain: the primary kingdoms|journal = Proc Natl Acad Sci USA|volume = 74 |pages = 5088-90 | year = 1977 | id = PMID 270744}}</ref>. The SSU rRNA as a measure of evolutionary distances was pioneered by [[Carl Woese]] when formulating the first modern "tree of life", and his results led him to propose the [[Archaea]] as a third domain of [[life]].) | *{{cite journal | author = Woese C, Fox G | title = Phylogenetic structure of the prokaryotic domain: the primary kingdoms|journal = Proc Natl Acad Sci USA|volume = 74 |pages = 5088-90 | year = 1977 | id = PMID 270744}}</ref>. The SSU rRNA as a measure of evolutionary distances was pioneered by [[Carl Woese]] when formulating the first modern "tree of life", and his results led him to propose the [[Archaea]] as a third domain of [[life]].) | ||
Microbiologists (led by [[Carl Woese]]) have introduced the term ''domain'' for the three main early branches of the tree of life, where ''domain'' is a [[phylogenetic]] term very similar in meaning to biological kingdom | Microbiologists (led by [[Carl Woese]]) have introduced the term ''domain'' for the three main early branches of the tree of life, where ''domain'' is a [[phylogenetic]] term very similar in meaning to biological kingdom. | ||
These three domains of life represent the main lineages in evolution of early cellular life and currently represented by the ''[[Bacteria]]'', the ''[[Archaea]]'' (single celled organisms superficially similar to bacteria), and ''[[Eukaryote|Eukarya]] (eukaryote)'' domains. Eukaryotes are all organisms with a well defined nucleus, and this domain comprises protists, fungi, and all organisms in the animal and plant kingdoms, including humans (See figure at left). | These three domains of life represent the main lineages in evolution of early cellular life and currently represented by the ''[[Bacteria]]'', the ''[[Archaea]]'' (single celled organisms superficially similar to bacteria), and ''[[Eukaryote|Eukarya]] (eukaryote)'' domains. Eukaryotes are all organisms with a well defined nucleus, and this domain comprises protists, fungi, and all organisms in the animal and plant kingdoms, including humans (See figure at left). | ||
As seen in the figure, studies of SSU rRNA genes (and some other genes) suggest that ''Archaea'' and ''Eukarya'' have a ''sister'' relationship in evolution, and it has often been assumed that eukaryotes evolved from archeal cells. | As seen in the figure, studies of SSU rRNA genes (and some other genes) might suggest that ''Archaea'' and ''Eukarya'' have a ''sister'' relationship in evolution, and it has often been assumed that eukaryotes evolved from archeal cells. | ||
But the fact that genes can move between distant branches of the tree of life even at low probabilities poses problems for scientists trying to reconstruct evolution from studying genes and gene sequences in different organisms, because HGT effectively scrambles the information which biologists rely on when reconstructing a phylogeny of organisms (i.e., their evolutionary history and relationships). The challenges that are raised by HGT are most awkward for the ambitious reconstruction of the earliest events in evolution - the early branches of the tree of life - because over a long enough time and with large numbers of organisms, many HGT events are certain to have occurred, despite the low probability of individual events. | But the fact that genes can move between distant branches of the tree of life even at low probabilities poses problems for scientists trying to reconstruct evolution from studying genes and gene sequences in different organisms, because HGT effectively scrambles the information which biologists rely on when reconstructing a phylogeny of organisms (i.e., their evolutionary history and relationships). The challenges that are raised by HGT are most awkward for the ambitious reconstruction of the earliest events in evolution - the early branches of the tree of life - because over a long enough time and with large numbers of organisms, many HGT events are certain to have occurred, despite the low probability of individual events. | ||
| Line 113: | Line 113: | ||
* Ge F ''et al'' (2005) The Cobweb of Life revealed by genome-scale estimates of horizontal gene transfer ''PLoS Biol'' [http://biology.plosjournals.org/perlserv/?request=get-document&doi=10.1371/journal.pbio.0030316 '''3'''(10):e316] | * Ge F ''et al'' (2005) The Cobweb of Life revealed by genome-scale estimates of horizontal gene transfer ''PLoS Biol'' [http://biology.plosjournals.org/perlserv/?request=get-document&doi=10.1371/journal.pbio.0030316 '''3'''(10):e316] | ||
*Fitzpatrick DA ''et al'' (2006) A fungal phylogeny based on 42 complete genomes derived from supertree and combined gene analysis ''BMC Evol Biol'' [http://www.biomedcentral.com/1471-2148/6/99 '''6''':99] | *Fitzpatrick DA ''et al'' (2006) A fungal phylogeny based on 42 complete genomes derived from supertree and combined gene analysis ''BMC Evol Biol'' [http://www.biomedcentral.com/1471-2148/6/99 '''6''':99] | ||
*[Steenkamp ET ''et al.''(2006) The protistan origins of animals and fungi. Mol Biol Evol. '''23'''(1):93-106. PMID 16151185] | |||
* Urwin R, Maiden MC (2003) Multi-locus sequence typing: a tool for global epidemiology ''Trends Microbiol'' '''11''':479-87</ref> | * Urwin R, Maiden MC (2003) Multi-locus sequence typing: a tool for global epidemiology ''Trends Microbiol'' '''11''':479-87</ref> | ||
Revision as of 00:42, 11 January 2007
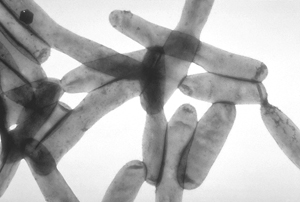
Horizontal gene transfer (HGT) (also called lateral gene transfer (LGT)) is any process in which an organism transfers genetic material to another cell or organism that is not one of its own offspring. HGT is thus very different from the normal vertical gene transfer whereby parental traits are inherited by the progeny, whether by sexual fusion of gametes to form zygotes as in animals and plants, or by asexual propagation as in microorganisms such as bacteria and fungi. HGT occurs at a much lower frequency than vertical gene transfer, so is not easily detected directly, and finding evidence for it requires special techniques.
Introduction
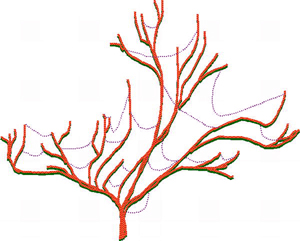
Advances in genome science and bioinformatics have brought abundant evidence that natural HGT has occurred between diverse biological taxa that are widely separated in the phylogenetic tree. Horizontal gene transfers include gene movement between different species of microbes and other microbial taxa such as protists, gene movement between different plant families, between different animals, and between bacteria and plants. Microorganisms are most affected by HGT, but recent research suggests that HGT in microbes is limited to about 2% of core genes in microbial genomes, and this is low enough to allow the the main lineages of microbial evolution to be treated as 'trees' with HGT 'cobwebs' (see figure to right).
Gene transfers between different biological sub-kingdoms (domains), such as between eukaryotic protists and bacteria [1] , or between bacteria and insects [2] are the most phylogenetically extreme cases of HGT. An example is bacterial "rol" genes from Agrobacterium species which have been found in tobacco plants (Nicotiniana). [3].
HGT is closely related to the dynamic changes that occur during genome evolution caused by the DNA rearrangement and transposition processes catalyzed by mobile DNA ("jumping genes", transposons). Movement of mobile genes (such as transposons) within a genome, and between different parts of an organism's genome (that is, between the chromosomes of the nucleus, the circular mitochondrion chromosome [4], and the circular plastid (chloroplast) chromosome) are part of the mechanisms that enable HGT between different species.
Main features of HGT in nature
- A hallmark of HGT is the presence of the same gene in organisms that are only very distantly related. The frequent discovery of shared DNA sequences such as the marinerclass of transposons, insertion sequence DNA, and retrovirus genes in diverse species, and shared mitochondrial genes in diverse flowering plants, indicate that mobile DNA has natural pathways for movement between species. Close relatives of mariner mobile DNA have been discovered in organisms as diverse as mites, flatworms, hydras, insects, nematodes, mammals and humans[5]. (The name mariner for this transposon an allusion to The Rhyme of the Ancient Mariner, meaning a traveller to distant lands.)

- Horizontal movement of genes is common among bacteria, and is a major facor in accelerating the rate of evolution in these organisms.[6]. It is involved in infectious multiple-antibiotic resistance in pathogenic bacteria, a major factor limiting the effectiveness of antibiotics. Inter-domain(sub-kingdom) transfer of several genes from eukaryotes to bacteria for instance, has occurred in the "accidentally pathogenic" bacterium (Legionella pneumophila, see illustration) that lives and replicates within a vacuole of protist and mammalian macrophage cells [7].
- HGT is common in diverse groups of unicellular protists, which often contain several genes transferred from both prokaryotes and other protists [8].
- HGT occurs globally on a massive scale among marine microorganisms, and viruses, at total numbers near 1029 being the most common biological entities in the sea, are a major pathway for inter-species gene movement in the ocean. The estimated virus-mediated gene transfer events in the Mediterranean sea are 1013 per year [9]. Endosymbiosis with an alga is identified as a route for HGT in marine dinoflagellates, the organisms that cause 'red tides' [10].
- Mechanisms for HGT in flowering plants involving parasitic plants such as dodder or endophytes such as mosses (which facilitate inter-species gene transfer by being in intimate cell-to-cell contact with their host plants) are now well established (see Horizontal gene transfer in plants).
- Not all of the vehicles by which HGT occurs are fully characterized, but some are clearly identified. HGT is difficult to detect directly, as it occurs at lower frequencies than with normal sexual reproduction within the species. Modern techniques of DNA analysis, by providing detailed comparison of genomes, provide much of the evidence for HGT. In insects, mites and viruses are probable vectors for HGT. In bacteria, surface appendages called pili have various roles in DNA uptake, DNA secretion and DNA transfer which have been extensively analyzed; HGT in bacteria includes plasmid-mediated promiscuous mating by bacteria, for instance by the crown-gall bacterium Agrobacterium tumefaciens[11], and carriage of genes between species by viruses[12]. Direct DNA uptake as another transfer mechanism is illustrated by Legionella bacteria, which are naturally competent for DNA uptake.
Prokaryotes
- See main article HGT in prokaryotes
- The three main mechanisms of HGT in bacteria and archaea discussed here are:
- Bacterial Transformation or direct uptake of extracellular DNA.
- Transduction of genes by bacterial viruses.
- Bacterial conjugation, a gene transfer process carried out by plasmids and conjugative transposons.
Eukaryotes
Protists
Analysis of the complete genome sequence of the protist Entamoeba histolytica indicates 96 cases of relatively recent HGT from prokaryotes [13], whereas similar analysis of the complete genome sequence of the protist Cryptosporidium parvum reveals 24 candidates of HGT from bacteria [14].There is also convincing evidence that a bacterial gene for a biosynthetic enzyme has been recruited by the protist Trichomonas vaginalis from bacteria related to the ancestors of Pasteurella bacteria.[15] These results fit the idea that "you are what you eat". That is, with unicellular grazing organisms, foreign genetic material is constantly entering the cell and occasionally the genome from food organisms [16]
Fungi
Comparison of the genome sequences of two fungi, baker's yeast (Saccharomyces cerevisiae) and Ashbya gossypii, has shown that Saccharomyces has received two genes from bacteria by HGT. One codes for an enzyme that allows baker's yeast to make pyrimidine nucleotide bases anaerobically, and the other allows usage of sulfur from several organic sulfur sources.[17]. Other work with yeasts suggests that eight genes from Yarrowia lipolytica, five from Kluyveromyces lactis, and one from Debaryomyces hansenii are horizontally transferred. [18]
Other eukaryotes
Analysis of DNA sequences suggests that HGT has also occurred within multicellular eukaryotes, by a route that involves transfer of genes from chloroplast and mitochondrial genomes to the nuclear genomes [19]. According to the endosymbiotic theory, chloroplasts and mitochondria originated as the bacterial endosymbionts of a progenitor to the eukaryotic cell.
Plants
- See Horizontal gene transfer in plants for
- Natural gene transfer between plants that do not cross-pollinate
- Jumping genes cross naturally between rice and millet
- Epiphytes and parasites as a bridge for gene flow between diverse plant species
- See Transgenic plant for hybridization by cross-pollination and artificial horizontal gene transfer in biotechnology.
Plant genes have also been discovered to be able to move to endophyte fungi that grow on them. Several plant endophyte fungi that grow on taxol-producing yew trees have gained the ability to make taxol themselves [20]. (Taxol, also called paclitaxel, is an anti-cancer drug found in yew trees.)
Animals
Junk DNA is the most obvious general evidence of HGT in eukaryotes. Such seemingly non-functional repetitive DNA is a major portion of many genomes of plants and animals. This DNA usually includes multiple copies of various "Jumping genes" which can proliferate within a genome after they have been transferred from another species. Examples in the human of such mobile elements are 'Hsmar1' and 'Hsmar2' which are related to the widely studied 'mariner' transposon. Close relatives of mariner mobile DNA have been discovered in organisms as diverse as mites, flatworms, hydras, insects, nematodes, mammals and humans [21].Retroviruses and retrotransposons are other examples of mobile horizontally transferred DNA found in animals.
The adzuki bean beetle Callosobruchus chinensis is infected with several strains of bacterial Wolbachia endosymbionts. A genome fragment of one of these endosymbionts has been found transferred to the X chromosome of the host insect [22].
History of discovery of HGT
- See main article Horizontal gene transfer (History)
- Bacterial genetics starts in 1946
- see main article Horizontal gene transfer in prokaryotes
- First glimpses of horizontal transfer of traits in plant evolution
- see also main article Barbara McClintock
- Discovery of mobile genes in flies, and mariner
- HGT and genetic engineering
Decoding the tree of life from genomes scrambled by HGT
Because each organism carries a record of its ancestry in its DNA, methods for rapid gene isolation and analysis of DNA encoded sequence - which extract information from this ancestral archive - have provided powerful tools for answering questions about evolution. These techniques have enabled rapid expansion of the field of biological systematics known as phylogenetic inference. Comparison of related (homologous) gene sequences ffrom different organisms has been widely and successfully applied to reconstruct the history of many evolutionary lineages. [23]
Numerous issues about evolution have been clarifed by comparing homologous genes from different species, genera, families, and phyla. Unfortunately HGT complicates the use of DNA sequences to reconstruct evolutionary history because HGT scrambles the genetic evidence needed to deduce the branching patterns of evolutionary trees.
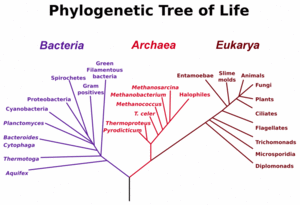
One area of current research in phylogenetic inference, and arguably one the most challenging problems in evolutionary theory, is the early stages in the evolution of life. This topic provides a good illustration of the complications introduced by HGT into the reconstruction of evolution.
The main early branches of the tree of life have been intensively studied by microbiologists because the first organisms were microorganisms.
A very common gene used for constructing phylogenetic relationships in microrganisms is the small ribosomal subunit ribosomal RNA gene (SSU rRNA) gene, as its sequences tend to be conserved among members with close phylogenetic distances, yet it is variable enough that differences can be measured [24]. The SSU rRNA as a measure of evolutionary distances was pioneered by Carl Woese when formulating the first modern "tree of life", and his results led him to propose the Archaea as a third domain of life.)
Microbiologists (led by Carl Woese) have introduced the term domain for the three main early branches of the tree of life, where domain is a phylogenetic term very similar in meaning to biological kingdom.
These three domains of life represent the main lineages in evolution of early cellular life and currently represented by the Bacteria, the Archaea (single celled organisms superficially similar to bacteria), and Eukarya (eukaryote) domains. Eukaryotes are all organisms with a well defined nucleus, and this domain comprises protists, fungi, and all organisms in the animal and plant kingdoms, including humans (See figure at left).
As seen in the figure, studies of SSU rRNA genes (and some other genes) might suggest that Archaea and Eukarya have a sister relationship in evolution, and it has often been assumed that eukaryotes evolved from archeal cells.
But the fact that genes can move between distant branches of the tree of life even at low probabilities poses problems for scientists trying to reconstruct evolution from studying genes and gene sequences in different organisms, because HGT effectively scrambles the information which biologists rely on when reconstructing a phylogeny of organisms (i.e., their evolutionary history and relationships). The challenges that are raised by HGT are most awkward for the ambitious reconstruction of the earliest events in evolution - the early branches of the tree of life - because over a long enough time and with large numbers of organisms, many HGT events are certain to have occurred, despite the low probability of individual events.
Recent discoveries of 'rampant' HGT in microorganisms, and the detection of horizontal movement of even genes for the small subunit of ribosomal RNA have forced biologists to question the accuracy of at least the early branches in the tree shown on the left, and even question the validity of trees as useful models of how microbial evolution occurs.[25] [26]
Recent efforts to infer evolutionary trees while recognizing HGT
Difficulties in acertaining correct branching structure for the evolutionary tree of microbes that are introduced because of occurence of HGT , and especially uncertainties the earliest branches in the tree of life, have been a great spur for current biological research. Clear answers to all questions about these trees are not yet available, but many interesting discoveries and ideas have surfaced because of this effort.
Instead of relying a single gene such as the SSU rRNA gene to reconstruct evolution, scientific effort has shifted to seeking evidence from the numerous now available complete genome sequences of organisms to obtain more reliable models for evolution.[27]
So far, this research endeavor suggests that most of the core genes of bacteria are unaffected by HGT, so that if evidence from many genes is used to investigate how lineages are related to one another, reliable tree can be deduced for a least the more recent stages in microbial evolution. Thus is argued that the 'tree' is still a valid metaphor for microbial evolution - but a tree is adorned with 'cobwebs' of horizontally transferred genes.
But it is also clear that the deep branches in trees based only on SSU rRNA do not capture all the events of early eukaryote evolution accurately. For instance, careful analysis of the complete genome of the eukaryote yeast shows that it contains many genes more that closely related to bacterial genes than to archaea, is is now apparent that archaea were not the simple progenitors of the eukaryotes, a contradiction of the findings with investigations based on SSU rRNA and a limited same of other genes. [28]
One interesting hypotheses supported by recent analysis of complete genome sequences is that the first nucleated cell arose as a genome fusion between two diverse prokayotes - one an archaen cell, and the other a true bacterium.
Such an genome fusion event may have occurred in ancient consortia of microbes that worked together to pool their metablic capabilities, and a gene fusion might have plausibly involved two prokaryote partners of a symbiosis. Several variatons of this symbiosis hypothesis have been suggested by different scientists.[29]
See also
- Gene flow
- Phylogenetic tree
- Endogenous retrovirus
- Germline
- Mitochondrion
- Integron
- Provirus
- Retrotransposon
- Mobile DNA
- Pilus
References
Citations
- ↑ Suwwan de Felipe K et al (2005) Evidence for acquisition of Legionella type IV secretion substrates via interdomain horizontal gene transfer J Bacteriol 187:7716-26
- ↑ Kondo N et al (2002) Genome fragment of Wolbachia endosymbiont transferred to X chromosome of host insect Proc Natl Acad Sci USA 99:14280-5
- ↑ Intrieri MC, Buiatti M (2001) The horizontal transfer of Agrobacterium rhizogenes genes and the evolution of the genus Nicotiana. Mol Phylogen Evol 20:100-10 PMID 11421651
- ↑ Adams KL et al(2000) Repeated, recent and diverse transfers of a mitochondrial gene to the nucleus in flowering plants Nature 408:354 PMID 11099041
- ↑ Robertson HM (1993) The mariner transposable element is widespread in insects Nature 362:241-5 PMID 8384700
- Robertson HM (1996) Reconstruction of the ancient mariners of humans Nature Genetics 12:360-1 PMID 8630486
- ↑ Jain R et al.(2003) Horizontal gene transfer accelerates genome innovation and evolution. Mol Biol Evol. 2003 20:1598-602. PMID 12777514
- ↑ de Felipe KS et al (2005) Evidence for acquisition of Legionella type IV secretion substrates via interdomain horizontal gene transfer J Bacteriol 187:7716-26
- ↑ Richards TA et al (2003) Protist 1:17–32 PMID 12812367
- Graham H et al (2003) The amitochondriate eukaryote Trichomonas vaginalis contains a divergent thioredoxin-linked peroxiredoxin antioxidant system JBC M304359200
- Andersson JO et al (2006) Evolution of four gene families with patchy phylogenetic distributions: influx of genes into protist genomes BMC Evol Biol 6:27
- de Koning AP et al (2000) Lateral gene transfer and metabolic adaptation in the human parasite Trichomonas vaginalis. Mol Biol Evol 17:1769-73
- Loftus B et al (2005) The genome of the protist parasite Entamoeba histolytica. Nature 433:865-8 PMID 15729342
- Huang J et al (2004) Phylogenomic evidence supports past endosymbiosis, intracellular and horizontal gene transfer in Cryptosporidium parvum. Genome Biology 5:R88
- ↑ Weinbauer MG et al (2004) Are viruses driving microbial diversification and diversity? Envir Microbiol 6:1-11
- Paul JH (1999) Microbial gene transfer J Mol Microbiol Biotechnol 1:45–50
- Fuhrman JA (1999) Marine viruses and their biogeochemical and ecological effects Nature 399:541–8 PMID 10376593
- ↑ Yoon HS et al (2005) Tertiary endosymbiosis driven genome evolution in dinoflagellate algae Mol Biol Evol 22:1299-308
- ↑ Zhu J et al (2000) The bases of crown gall tumorigenesis J Bacteriol 182:3885-95 This article describes the biology of crown-gall bacterium, and the mechanism of DNA injection by this bacterium, and explains how genes can move between bacterial species and from bacteria to eukaryotic organisms, and illustrates the extent to which different species can co-evolve
- ↑ Weinbauer et al (2004) Are viruses driving microbial diversification and diversity? Envir Microbiol 6:1-11
- Amoils S (2005) Analysing incompatibility — Wolbachia on the couch Nature Rev Microbiol 3:667
- Besser TE et al (2006) Greater diversity of Shiga toxin-encoding bacteriophage insertion sites among Escherichia coli O157:H7 isolates from cattle than from humans Appl Environ Microbiol PMID 17142358
- ↑ Loftus B et al (2005) The genome of the protist parasite Entamoeba histolytica. Nature 433:865-8 PMID 15729342
- ↑ Huang J et al (2004) Phylogenomic evidence supports past endosymbiosis, intracellular and horizontal gene transfer in Cryptosporidium parvum. Genome Biology 5:R88 PMID 15535864
- ↑ de Koning et al (2000) Lateral gene transfer and metabolic adaptation in the human parasite Trichomonas vaginalis. Mol Biol Evol 17:1769-73
- ↑ Doolittle WF (1998) You are what you eat: a gene transfer ratchet could account for bacterial genes in eukaryotic nuclear genomes Trends in Genetics 14:307-11 PMID 9724962
- ↑ Hall CS et al(2005) Contribution of horizontal gene transfer to the evolution of Saccharomyces cerevisiae. Eukaryot Cell 4:1102-1115
- ↑ Dujon B et al {2004) Genome evolution in yeasts Nature 430:35-44 PMID 15229592
- ↑ Gray MW (1993) Origin and evolution of organelle genomes Curr Opin Genet Dev 3:884-90 PMID 8118213
- ↑ Shrestha K et al (2001) Evidence for paclitaxel from three new endophytic fungi of Himalayan yew of Nepal Planta Med 67:374-6 PMID 11458463
- ↑ Robertson HM et al (1996) Reconstruction of the ancient 'mariners' of humans Nature Genetics 12:360-361 PMID 8630486
- ↑ Kondo N et al (2002) Genome fragment of Wolbachia endosymbiont transferred to X chromosome of host insect Proc Natl Acad Sci USA 99:14280-5
- ↑ Adoutte A et al. (2000) The new animal phylogeny: reliability and implications. Proc Natl Acad Sci U S A. 97(9):4453-6. PMID 10781043
- [Steenkamp ET et al.(2006) The protistan origins of animals and fungi. Mol Biol Evol. 23(1):93-106. PMID 16151185]
- ↑ Woese C et al (1990). "Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya". Proc Natl Acad Sci USA 87: 4576-9. PMID 2112744.
- Woese C, Fox G (1977). "Phylogenetic structure of the prokaryotic domain: the primary kingdoms". Proc Natl Acad Sci USA 74: 5088-90. PMID 270744.
- ↑ Simonson AB et al (2005) Decoding the genomic tree of life Proc Natl Acad Sci USA 102 Suppl 1:6608-13. PMID 15851667
- ↑ Yap WH et al (1999) Distinct types of rRNA operons exist in the genome of the actinomycete Thermomonspora chromogena and evidence for horizontal gene transfer of an entire rRNA operon J Bacteriol 181:5201-9 PMID 10464188
- ↑ Eisen JA, Fraser CM (2003) Viewpoint phylogenomics: intersection of evolution and genomics Science 300:1706-7 DOI: 10.1126/science.1086292
- Henz SR et al (2005) Whole-genome prokaryotic phylogeny Bioinformatics 21:2329-35 PMID 15166018
- Ge F et al (2005) The Cobweb of Life revealed by genome-scale estimates of horizontal gene transfer PLoS Biol 3(10):e316
- Fitzpatrick DA et al (2006) A fungal phylogeny based on 42 complete genomes derived from supertree and combined gene analysis BMC Evol Biol 6:99
- [Steenkamp ET et al.(2006) The protistan origins of animals and fungi. Mol Biol Evol. 23(1):93-106. PMID 16151185]
- Urwin R, Maiden MC (2003) Multi-locus sequence typing: a tool for global epidemiology Trends Microbiol 11:479-87
- ↑ Esser, C et al. (2004) A Genome Phylogeny for Mitochondria Among alpha-Proteobacteria and a preedominantly eubacterial ancestry of yeast nuclear genes. Mol. Biol. Evol. 21:1643-50. PMID 15155797
- Rivera MC and Lake JA (2004) The ring of life provides evidence for a genome fusion origin of eukaryotes. Nature 431:152-5 PMID 15356622
- Simonson AB et al. Decoding the genomic tree of life. (2005) Proc Natl Acad Sci U S A. 102 Suppl 1:6608-13. PMID 15851667
- ↑ Esser, C et al. (2004) A Genome Phylogeny for Mitochondria Among alpha-Proteobacteria and a preedominantly eubacterial ancestry of yeast nuclear genes. Mol. Biol. Evol. 21:1643-50. PMID 15155797
Further Reading
- Horizontal Gene Transfer Syvanen M, Kado CI (2002) 2nd edition, Academic Press ISBN 0-12-680126-6 A comprehensive treatise. Reviewed here by M-W Ho
- Acquiring genomes: a theory of the origin of species. Margulis L and Sagan D (2002) Basic Books ISBN 0-465-04392-5. A book that looks at gene transfer from a different perspective to many conventional interpretations with an interesting emphasise on microbial diversity. Reviewed here.
- Mobile DNA. Berg DE, Howe MM (Eds.)(1989) American Society for Microbiology. Washington D.C. Book with a comprehensive discussion of mobile DNA, jumping genes, transposons etc in many organisms, not only bacteria. Reviewed by Freeling M (1990) in Q Rev Biol 65:217-8
- Salzberg SL et al (2001) Microbial genes in the human genome: lateral transfer or gene loss?" Science 292:1903-6. This reports that one dramatic claim of HGT - in which a distinguished group of scientists claimed that bacteria transferred their DNA directly into the human lineage - was simply wrong [1]
- Weinbauer MG, Rassoulzadegan F (2004) Are viruses driving microbial diversification and diversity? Envir Microbiol 6:1-11 Discussion of both the evolutionary and ecological activities of viruses in the ocean, a major source of HGT in nature.
- Woese C (2002) On the evolution of cells Proc Natl Acad Sci USA99:8742-7 This article shifts the emphasis in early phylogenic adaptation from vertical to horizontal gene transfer.
- Hall C et al (2005) Contribution of horizontal gene transfer to the evolution of Saccharomyces cerevisiae Eukaryot Cell 4:1102-15Convincing evidence of horizontal transfer of bacterial DNA:
- Snel B et al (1999) Genome phylogeny based on gene content Nature Genetics 21:66-7 [2]Proposal for using the presence or absence of a set of genes to infer phylogenies, in order to avoid confounding factors such as HGT.
External links
- Webfocus in Nature with free access review articles Focus on horizontal gene transfer
- (1999) Horizontal gene transfer among genomes: The complexity hypothesis Proc Natl Acad Sci USA 96:3801-6
- Ochman H et al (2000) Lateral gene transfer and the nature of bacterial innovation (pdf)
- The New Yorker July 12, 1999, p44-61 "Smallpox knows how to make a mouse protein. How did smallpox learn that? 'The poxviruses are promiscuous at capturing genes from their hosts,' Esposito said. 'It tells you that smallpox was once inside a mouse or some other small rodent'"
- Retrotransfer or gene capture: a feature of conjugative plasmids, with ecological and evolutionary significance
- Horizontal gene transfer - A new paradigm for biology
- Horizontal gene transfer (p334 of Molecular Genetics by Ulrich Melcher)
- Report on horizontal gene transfer by Mae-Wan Ho, 1999
- Recent evidence confirms risks of horizontal gene transfer
- Horizontal gene transfer at sciences.sdsu.edu
